Running SCENIC+#
This tutorial will illustrate how to run the SCENIC+ pipeline via Snakemake.
Before running this pipeline you should:
Preprocess the scATAC-seq side of the data using pycisTopic, click here for a tutorial.
Preprocess the scRNA-seq side of the data using Scanpy, click here for a tutorial.
Optionally, but highly recommended, generate a cisTarget database using the consensus peaks specific to your dataset, click here for a tutorial.
In case you have human, mouse or fly data you can also use one of the precomputed cisTarget databases. These can be found on our resources website.
[1]:
import os
os.chdir("/staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial")
SCENIC+ can be run entirely using the command line. Please refer to the docummentation (by using the --help flag) for more information.
Below we will use the Snakemake pipeline, which is already included in SCENIC+ to perform the analysis.
[3]:
!scenicplus
____ ____ _____ _ _ ___ ____
/ ___| / ___| ____| \ | |_ _/ ___| _
\___ \| | | _| | \| || | | _|.|_
___) | |___| |___| |\ || | |__|_..._|
|____/ \____|_____|_| \_|___\____||_|
scenicplus verions: 1.0a1
usage: scenicplus [-h] {init_snakemake,prepare_data,grn_inference} ...
Single-Cell Enhancer-driven gene regulatory Network Inference and Clustering
positional arguments:
{init_snakemake,prepare_data,grn_inference}
options:
-h, --help show this help message and exit
scRNA-seq preparation#
scRNA-seq side of the experiment can be processed according to the regular Scanpy tutorial. Just make sure to store the raw gene expresison matrix in adata.raw.
Call the following piece of code:
adata.raw = adata
BEFORE normalizing the data
sc.pp.normalize_total(adata, target_sum=1e4)
sc.pp.log1p(adata)
[41]:
ls data
10x_multiome_brain_cisTopicObject_noDBL.pkl@ adata.h5ad@ region_sets/
Initialize Snakemake#
To run Snakemake we first have to initialize the pipeline. This will create a folder named Snakemake containing a folder for the config.yaml file and a folder containing the actual workflow definition.
[45]:
!mkdir -p scplus_pipeline
!scenicplus init_snakemake --out_dir scplus_pipeline
2024-03-06 14:28:10,766 SCENIC+ INFO Creating snakemake folder in: scplus_pipeline
[51]:
!tree scplus_pipeline/
scplus_pipeline/
└── Snakemake
├── config
│ └── config.yaml
└── workflow
└── Snakefile
3 directories, 2 files
[2]:
!mkdir -p outs
!mkdir -p tmp
Next, modify the config.yaml file.
The most important fields are input_data and output_data. For the other default values can be kept:
input_data: This is the input to the pipeline, these files should already exist.
cisTopic_obj_fname: the path to your cistopic object containing processed chromatin accessibility data.GEX_anndata_fname: the path to your scanpy h5ad file containing processed gene expression data.region_set_folder: the path to directory containing several directories with bed files. Differential motif enrichment (1 vs all) will be run within each sub folder. As an example the structure of the folder is shown below.ctx_db_fname: the path to the cisTarget ranking database.dem_db_fname: the path to the cisTarget score database.path_to_motif_annotations: the path to the motif-to-TF annotaiton. For human (hgnc), mouse (mgi), chicken and fly (flybase) these files can be downloaded from our resources website, please download the relevant file starting with “motifs-v10nr_clust”.
output_data: This is the output of the pipeline, these files will be created. If some of these files already exists (for example when the pipeline has only been partially run) some steps of the workflow might be skipped.
combined_GEX_ACC_mudata: where the MuData object containing gene expression and imputed chromatin accessibility should be stored.dem_result_fname: where the h5 file containing DEM based enriched motifs should be stored.ctx_result_fname: where the h5 file containing cistarget based enriched motifs should be stored.output_fname_dem_html: where the html file containing DEM based enriched motifs should be stored.output_fname_ctx_html: where the html file containing cistarget based enriched motifs should be stored.cistromes_direct: where the AnnData h5ad file should be stored containing TF-to-region links based on direct motif-to-TF annotations.cistromes_extended: where the AnnData h5ad file should be stored containing TF-to-region links based on exteded (e.g. orthology based) motif-to-TF annotations.tf_names: where a text file containing TF names, based on the enriched motifs, should be stored.genome_annotation: where a data frame (tsv) should be stored containing genome annotation.chromsizes: where the chromsizes file should be stored.search_space: where the search space for each gene should be stored.tf_to_gene_adjacencies: where the TF-to-gene links, with importance scores, should be stored.region_to_gene_adjacencies: where the region-to-gene links, with importance scores, should be stored.eRegulons_direct: where the dataframe (tsv) containing eRegulons (TF-region-gene links) based on direct motif-to-TF annotations should be stored.eRegulons_extended: where the dataframe (tsv) containing eRegulons (TF-region-gene links) based on extended (e.g. orthology based) motif-to-TF annotations should be stored.AUCell_direct: where the MuData containing target gene and target region enrichement scores for each cells, based on direct motif-to-TF annotations should be stored.AUCell_extended: where the MuData containing target gene and target region enrichement scores for each cells, based on extended (e.g. orthology based) motif-to-TF annotations should be stored.scplus_mdata: where the final output MuData containing AUCell values and the (TF-region-gene links) based on both direct and extended motif-to-TF annotations should be stored.
params_general General parameters. - temp_dir: Directory to store temporary data. - n_cpu: maximum number of CPU’s to use. - seed: seed to use to initialize the random state.
params_data_preparation Parameters used for the data preparation step. - bc_transform_func: lambda function to transform the scRNA-seq barcode so they match with the scATAC-seq ones - is_multiome: boolean specifying wether the data is multiome or not. - key_to_group_by: in case of non-multiome data, cell metadata variable to group cells by in order to generate metacells that can be matched across the scRNA-seq and scATAC-seq side of the data. This variable should be prefixed with
eiter “GEX:” or “ACC:”. - nr_cells_per_metacells: in case of non-multione data, number of cells to sample to sample for each metacell. - direct_annotation: Which annotations fields to use for generating direct motif-to-TF annotations. - extended_annotation: Which annotations fields to use for generating extended motif-to-TF annotations - species: Species name, for example “hsapiens” - biomart_host: Biomart host to use for downloading genome annotations. Make sure that this
host matches the genome reference you are using, please visit this website for more information. - search_space_upstream: string in the form “<minmal> <maximal>” specifying the <minimal> and <maximal> search space to consider downstream of the TSS of each gene. - search_space_downstream: string in the form “<minmal> <maximal>” specifying the <minimal> and <maximal> search space to consider upstrean of the TSS of each gene. -
search_space_extend_tss: string in the form “<upstream> <downstream>” specifying the amount of basepairs the TSS of each gene should be extended, <upstream> and
params_motif_enrichment parameters for performing motif enrichment analysis. - species: Species used for the analysis. This parameter is used to download the correct motif-to-TF annotations from the cisTarget webservers. - annotation_version: Version of the motif-to-TF annotation to use. This parameter is used to download the correct motif-to-TF data from the cisTarget webservers. - motif_similarity_fdr: Threshold on motif similarity scores for calling similar motifs. -
orthologous_identity_threshold: Threshold on the protein-protein orthology score for calling orthologous motifs - annotations_to_use: Which annotations to use for annotation motifs to TFs. - fraction_overlap_w_dem_database: Fraction of nucleotides, of regions in the bed file, that should overlap with regions in the scores database. - dem_max_bg_regions: Maximum number of regions to use as background for DEM. - dem_balance_number_of_promoters: Boolean specifying wether the
number of promoters should be equalized between the foreground and background set of regions. - dem_promoter_space: Number of basepairs up- and downstream of the TSS that are considered as being the promoter for that gene. - dem_adj_pval_thr: Threshold on the Benjamini-Hochberg adjusted p-value from the Wilcoxon test performed on the motif score of foreground vs background regions for a motif to be considered as enriched. - dem_log2fc_thr: Threshold on the log2 fold change of the
motif score of foreground vs background regions for a motif to be considered as enriched. - dem_mean_fg_thr: Minimul mean signal in the foreground to consider a motif enriched for DEM. - dem_motif_hit_thr: Minimal CRM score to consider a region enriched for a motif for DEM. - fraction_overlap_w_ctx_database: Fraction of nucleotides, of regions in the bed file, that should overlap with regions in the ranking database. - ctx_auc_threshold: Threshold on the AUC value for calling
significant motifs - ctx_nes_threshold: Threshold on the NES value for calling significant motifs. - ctx_rank_threshold: The total number of ranked regions to take into account when creating a recovery curves.
params_inference Parameters for performing GRN inference. - tf_to_gene_importance_method: Method to use to calculate TF-to-gene importance scores. - region_to_gene_importance_method: Method to use to calculate region-to-gene importance scores. - region_to_gene_correlation_method: Method to use to calculate region-to-gene correlation coefficients. - order_regions_to_genes_by: Value to order region-to-gene scores by for selecting top regions per gene -
order_TFs_to_genes_by: value to order TF-to-gene scores by for selecting top TFs per gene - gsea_n_perm: Number or permutations to perform for calculating GSEA enrichment scores. - quantile_thresholds_region_to_gene: space seperated list containing quantile threshold to be used for binarizing region-to-gene links. - top_n_regionTogenes_per_gene: space seperated list containing the number of top regions per gene for binarizing region-to-gene links. -
top_n_regionTogenes_per_region: space seperated list containging per region the number of top genes for binarizing region-to-gene links - min_regions_per_gene: minimum number of regions per gene for the link to be included in eGRNs. - rho_threshold: absolute threshold on the correlation coefficient to seperate positive and negative region-to-gene and TF-to-gene links - min_target_genes: minimum number of target genes per TF for the link(s) to be includedin eGRNs.
[1]:
!tree /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/pycisTopic_polars_tutorial/outs/region_sets
/staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/pycisTopic_polars_tutorial/outs/region_sets
├── DARs_cell_type
│ ├── AST.bed
│ ├── BG.bed
│ ├── COP.bed
│ ├── ENDO.bed
│ ├── GC.bed
│ ├── GP.bed
│ ├── INH_PVALB.bed
│ ├── INH_SNCG.bed
│ ├── INH_SST.bed
│ ├── INH_VIP.bed
│ ├── MG.bed
│ ├── MGL.bed
│ ├── MOL.bed
│ ├── NFOL.bed
│ ├── OPC.bed
│ └── PURK.bed
├── Topics_otsu
│ ├── Topic1.bed
│ ├── Topic10.bed
│ ├── Topic11.bed
│ ├── Topic12.bed
│ ├── Topic13.bed
│ ├── Topic14.bed
│ ├── Topic15.bed
│ ├── Topic16.bed
│ ├── Topic17.bed
│ ├── Topic18.bed
│ ├── Topic19.bed
│ ├── Topic2.bed
│ ├── Topic20.bed
│ ├── Topic21.bed
│ ├── Topic22.bed
│ ├── Topic23.bed
│ ├── Topic24.bed
│ ├── Topic25.bed
│ ├── Topic26.bed
│ ├── Topic27.bed
│ ├── Topic28.bed
│ ├── Topic29.bed
│ ├── Topic3.bed
│ ├── Topic30.bed
│ ├── Topic31.bed
│ ├── Topic32.bed
│ ├── Topic33.bed
│ ├── Topic34.bed
│ ├── Topic35.bed
│ ├── Topic36.bed
│ ├── Topic37.bed
│ ├── Topic38.bed
│ ├── Topic39.bed
│ ├── Topic4.bed
│ ├── Topic40.bed
│ ├── Topic5.bed
│ ├── Topic6.bed
│ ├── Topic7.bed
│ ├── Topic8.bed
│ └── Topic9.bed
└── Topics_top_3k
├── Topic1.bed
├── Topic10.bed
├── Topic11.bed
├── Topic12.bed
├── Topic13.bed
├── Topic14.bed
├── Topic15.bed
├── Topic16.bed
├── Topic17.bed
├── Topic18.bed
├── Topic19.bed
├── Topic2.bed
├── Topic20.bed
├── Topic21.bed
├── Topic22.bed
├── Topic23.bed
├── Topic24.bed
├── Topic25.bed
├── Topic26.bed
├── Topic27.bed
├── Topic28.bed
├── Topic29.bed
├── Topic3.bed
├── Topic30.bed
├── Topic31.bed
├── Topic32.bed
├── Topic33.bed
├── Topic34.bed
├── Topic35.bed
├── Topic36.bed
├── Topic37.bed
├── Topic38.bed
├── Topic39.bed
├── Topic4.bed
├── Topic40.bed
├── Topic5.bed
├── Topic6.bed
├── Topic7.bed
├── Topic8.bed
└── Topic9.bed
3 directories, 96 files
[122]:
!bat scplus_pipeline/Snakemake/config/config.yaml
───────┬────────────────────────────────────────────────────────────────────────
│ File: scplus_pipeline/Snakemake/config/config.yaml
───────┼────────────────────────────────────────────────────────────────────────
1 │ input_data:
2 │ cisTopic_obj_fname: "/staging/leuven/stg_00002/lcb/sdewin/PhD/python_
│ modules/pycisTopic_polars_tutorial/outs/cistopic_obj.pkl"
3 │ GEX_anndata_fname: "/staging/leuven/stg_00002/lcb/sdewin/PhD/python_m
│ odules/scenicplus_development_tutorial/data/adata.h5ad"
4 │ region_set_folder: "/staging/leuven/stg_00002/lcb/sdewin/PhD/python_m
│ odules/pycisTopic_polars_tutorial/outs/region_sets"
5 │ ctx_db_fname: "/staging/leuven/stg_00002/lcb/sdewin/PhD/python_module
│ s/scenicplus_development_tutorial/ctx_db/10x_brain_1kb_bg_with_mask.reg
│ ions_vs_motifs.rankings.feather"
6 │ dem_db_fname: "/staging/leuven/stg_00002/lcb/sdewin/PhD/python_module
│ s/scenicplus_development_tutorial/ctx_db/10x_brain_1kb_bg_with_mask.reg
│ ions_vs_motifs.scores.feather"
7 │ path_to_motif_annotations: "/staging/leuven/stg_00002/lcb/sdewin/PhD/
│ python_modules/scenicplus_development_tutorial/ctx_db/aertslab_motif_co
│ lleciton/v10nr_clust_public/snapshots/motifs-v10-nr.hgnc-m0.00001-o0.0.
│ tbl"
8 │
9 │ output_data:
10 │ # output for prepare_GEX_ACC .h5mu
11 │ combined_GEX_ACC_mudata: "/staging/leuven/stg_00002/lcb/sdewin/PhD/py
│ thon_modules/scenicplus_development_tutorial/outs/ACC_GEX.h5mu"
12 │ # output for motif enrichment results .hdf5
13 │ dem_result_fname: "/staging/leuven/stg_00002/lcb/sdewin/PhD/python_mo
│ dules/scenicplus_development_tutorial/outs/dem_results.hdf5"
14 │ ctx_result_fname: "/staging/leuven/stg_00002/lcb/sdewin/PhD/python_mo
│ dules/scenicplus_development_tutorial/outs/ctx_results.hdf5"
15 │ # output html for motif enrichment results .html
16 │ output_fname_dem_html: "/staging/leuven/stg_00002/lcb/sdewin/PhD/pyth
│ on_modules/scenicplus_development_tutorial/outs/dem_results.html"
17 │ output_fname_ctx_html: "/staging/leuven/stg_00002/lcb/sdewin/PhD/pyth
│ on_modules/scenicplus_development_tutorial/outs/ctx_results.html"
18 │ # output for prepare_menr .h5ad
19 │ cistromes_direct: "/staging/leuven/stg_00002/lcb/sdewin/PhD/python_mo
│ dules/scenicplus_development_tutorial/outs/cistromes_direct.h5ad"
20 │ cistromes_extended: "/staging/leuven/stg_00002/lcb/sdewin/PhD/python_
│ modules/scenicplus_development_tutorial/outs/cistromes_extended.h5ad"
21 │ # output tf names .txt
22 │ tf_names: "/staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/sc
│ enicplus_development_tutorial/outs/tf_names.txt"
23 │ # output for download_genome_annotations .tsv
24 │ genome_annotation: "/staging/leuven/stg_00002/lcb/sdewin/PhD/python_m
│ odules/scenicplus_development_tutorial/outs/genome_annotation.tsv"
25 │ chromsizes: "/staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/
│ scenicplus_development_tutorial/outs/chromsizes.tsv"
26 │ # output for search_space .tsb
27 │ search_space: "/staging/leuven/stg_00002/lcb/sdewin/PhD/python_module
│ s/scenicplus_development_tutorial/outs/search_space.tsv"
28 │ # output tf_to_gene .tsv
29 │ tf_to_gene_adjacencies: "/staging/leuven/stg_00002/lcb/sdewin/PhD/pyt
│ hon_modules/scenicplus_development_tutorial/outs/tf_to_gene_adj.tsv"
30 │ # output region_to_gene .tsv
31 │ region_to_gene_adjacencies: "/staging/leuven/stg_00002/lcb/sdewin/PhD
│ /python_modules/scenicplus_development_tutorial/outs/region_to_gene_adj
│ .tsv"
32 │ # output eGRN .tsv
33 │ eRegulons_direct: "/staging/leuven/stg_00002/lcb/sdewin/PhD/python_mo
│ dules/scenicplus_development_tutorial/outs/eRegulon_direct.tsv"
34 │ eRegulons_extended: "/staging/leuven/stg_00002/lcb/sdewin/PhD/python_
│ modules/scenicplus_development_tutorial/outs/eRegulons_extended.tsv"
35 │ # output AUCell .h5mu
36 │ AUCell_direct: "/staging/leuven/stg_00002/lcb/sdewin/PhD/python_modul
│ es/scenicplus_development_tutorial/outs/AUCell_direct.h5mu"
37 │ AUCell_extended: "/staging/leuven/stg_00002/lcb/sdewin/PhD/python_mod
│ ules/scenicplus_development_tutorial/outs/AUCell_extended.h5mu"
38 │ # output scplus mudata .h5mu
39 │ scplus_mdata: "/staging/leuven/stg_00002/lcb/sdewin/PhD/python_module
│ s/scenicplus_development_tutorial/outs/scplusmdata.h5mu"
40 │
41 │ params_general:
42 │ temp_dir: "/staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/sc
│ enicplus_development_tutorial/tmp"
43 │ n_cpu: 20
44 │ seed: 666
45 │
46 │ params_data_preparation:
47 │ # Params for prepare_GEX_ACC
48 │ bc_transform_func: "\"lambda x: f'{x}-10x_multiome_brain'\""
49 │ is_multiome: True
50 │ key_to_group_by: ""
51 │ nr_cells_per_metacells: 10
52 │ # Params for prepare_menr
53 │ direct_annotation: "Direct_annot"
54 │ extended_annotation: "Orthology_annot"
55 │ # Params for download_genome_annotations
56 │ species: "hsapiens"
57 │ biomart_host: "http://ensembl.org/"
58 │ # Params for search_space
59 │ search_space_upstream: "1000 150000"
60 │ search_space_downstream: "1000 150000"
61 │ search_space_extend_tss: "10 10"
62 │
63 │ params_motif_enrichment:
64 │ species: "homo_sapiens"
65 │ annotation_version: "v10nr_clust"
66 │ motif_similarity_fdr: 0.001
67 │ orthologous_identity_threshold: 0.0
68 │ annotations_to_use: "Direct_annot Orthology_annot"
69 │ fraction_overlap_w_dem_database: 0.4
70 │ dem_max_bg_regions: 500
71 │ dem_balance_number_of_promoters: True
72 │ dem_promoter_space: 1_000
73 │ dem_adj_pval_thr: 0.05
74 │ dem_log2fc_thr: 1.0
75 │ dem_mean_fg_thr: 0.0
76 │ dem_motif_hit_thr: 3.0
77 │ fraction_overlap_w_ctx_database: 0.4
78 │ ctx_auc_threshold: 0.005
79 │ ctx_nes_threshold: 3.0
80 │ ctx_rank_threshold: 0.05
81 │
82 │
83 │
84 │
85 │ params_inference:
86 │ # Params for tf_to_gene
87 │ tf_to_gene_importance_method: "GBM"
88 │ # Params regions_to_gene
89 │ region_to_gene_importance_method: "GBM"
90 │ region_to_gene_correlation_method: "SR"
91 │ # Params for eGRN inference
92 │ order_regions_to_genes_by: "importance"
93 │ order_TFs_to_genes_by: "importance"
94 │ gsea_n_perm: 1000
95 │ quantile_thresholds_region_to_gene: "0.85 0.90 0.95"
96 │ top_n_regionTogenes_per_gene: "5 10 15"
97 │ top_n_regionTogenes_per_region: ""
98 │ min_regions_per_gene: 0
99 │ rho_threshold: 0.05
100 │ min_target_genes: 10
101 │
───────┴────────────────────────────────────────────────────────────────────────
Run pipeline#
Once the config file is filled in the pipeline can be run.
[7]:
cd scplus_pipeline/Snakemake/
[8]:
ls
config workflow
[14]:
source /staging/leuven/stg_00002/mambaforge/vsc33053/etc/profile.d/conda.sh
conda activate scenicplus_development_tutorial
snakemake --cores 20
Assuming unrestricted shared filesystem usage for local execution.
Building DAG of jobs...
Using shell: /usr/bin/bash
Provided cores: 20
Rules claiming more threads will be scaled down.
Job stats:
job count
--------------------------- -------
AUCell_direct 1
AUCell_extended 1
all 1
download_genome_annotations 1
eGRN_direct 1
eGRN_extended 1
get_search_space 1
motif_enrichment_cistarget 1
motif_enrichment_dem 1
prepare_menr 1
region_to_gene 1
scplus_mudata 1
tf_to_gene 1
total 13
Select jobs to execute...
Execute 1 jobs...
[Mon Mar 11 15:16:45 2024]
localrule motif_enrichment_cistarget:
input: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/data/region_sets, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/ctx_db/10x_brain_1kb_bg_with_mask.regions_vs_motifs.rankings.feather, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/ctx_db/aertslab_motif_colleciton/v10nr_clust_public/snapshots/motifs-v10-nr.hgnc-m0.00001-o0.0.tbl
output: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/ctx_results.hdf5, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/ctx_results.html
jobid: 9
reason: Set of input files has changed since last execution; Params have changed since last execution
threads: 20
resources: tmpdir=/tmp
2024-03-11 15:16:50,142 SCENIC+ INFO Reading region sets from: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/data/region_sets
2024-03-11 15:16:50,142 SCENIC+ INFO Reading all .bed files in: topics_otsu
2024-03-11 15:16:50,367 SCENIC+ INFO Reading all .bed files in: topics_top_3k
2024-03-11 15:16:50,735 SCENIC+ INFO Reading all .bed files in: DARs_cell_type
2024-03-11 15:16:57,977 cisTarget INFO Reading cisTarget database
2024-03-11 15:16:58,101 cisTarget INFO Reading cisTarget database
2024-03-11 15:16:58,123 cisTarget INFO Reading cisTarget database
2024-03-11 15:16:58,184 cisTarget INFO Reading cisTarget database
2024-03-11 15:16:58,198 cisTarget INFO Reading cisTarget database
2024-03-11 15:16:58,203 cisTarget INFO Reading cisTarget database
2024-03-11 15:16:58,234 cisTarget INFO Reading cisTarget database
2024-03-11 15:16:58,273 cisTarget INFO Reading cisTarget database
2024-03-11 15:16:58,363 cisTarget INFO Reading cisTarget database
2024-03-11 15:16:58,369 cisTarget INFO Reading cisTarget database
2024-03-11 15:16:58,422 cisTarget INFO Reading cisTarget database
2024-03-11 15:16:58,426 cisTarget INFO Reading cisTarget database
2024-03-11 15:16:58,435 cisTarget INFO Reading cisTarget database
2024-03-11 15:16:58,519 cisTarget INFO Reading cisTarget database
2024-03-11 15:16:58,528 cisTarget INFO Reading cisTarget database
2024-03-11 15:16:58,537 cisTarget INFO Reading cisTarget database
2024-03-11 15:16:58,538 cisTarget INFO Reading cisTarget database
2024-03-11 15:16:58,763 cisTarget INFO Reading cisTarget database
2024-03-11 15:16:58,802 cisTarget INFO Reading cisTarget database
2024-03-11 15:16:58,842 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:05,150 cisTarget INFO Running cisTarget for topics_otsu_Topic31 which has 2449 regions
2024-03-11 15:17:05,978 cisTarget INFO Running cisTarget for topics_otsu_Topic11 which has 1374 regions
2024-03-11 15:17:06,523 cisTarget INFO Running cisTarget for topics_otsu_Topic35 which has 1590 regions
2024-03-11 15:17:06,712 cisTarget INFO Running cisTarget for topics_otsu_Topic5 which has 2008 regions
2024-03-11 15:17:06,773 cisTarget INFO Running cisTarget for topics_otsu_Topic12 which has 1443 regions
2024-03-11 15:17:07,062 cisTarget INFO Running cisTarget for topics_otsu_Topic37 which has 3458 regions
2024-03-11 15:17:07,317 cisTarget INFO Running cisTarget for topics_otsu_Topic17 which has 3147 regions
2024-03-11 15:17:07,410 cisTarget INFO Running cisTarget for topics_otsu_Topic26 which has 3691 regions
2024-03-11 15:17:07,564 cisTarget INFO Running cisTarget for topics_otsu_Topic25 which has 3108 regions
2024-03-11 15:17:07,721 cisTarget INFO Running cisTarget for topics_otsu_Topic9 which has 3601 regions
2024-03-11 15:17:07,825 cisTarget INFO Running cisTarget for topics_otsu_Topic33 which has 4570 regions
2024-03-11 15:17:08,573 cisTarget INFO Running cisTarget for topics_otsu_Topic2 which has 7021 regions
2024-03-11 15:17:08,656 cisTarget INFO Running cisTarget for topics_otsu_Topic28 which has 6562 regions
2024-03-11 15:17:08,765 cisTarget INFO Running cisTarget for topics_otsu_Topic30 which has 5719 regions
2024-03-11 15:17:08,772 cisTarget INFO Running cisTarget for topics_otsu_Topic40 which has 8153 regions
2024-03-11 15:17:08,954 cisTarget INFO Running cisTarget for topics_otsu_Topic15 which has 9880 regions
2024-03-11 15:17:09,141 cisTarget INFO Running cisTarget for topics_otsu_Topic38 which has 12221 regions
2024-03-11 15:17:09,457 cisTarget INFO Running cisTarget for topics_otsu_Topic29 which has 9544 regions
2024-03-11 15:17:09,678 cisTarget INFO Running cisTarget for topics_otsu_Topic1 which has 11489 regions
2024-03-11 15:17:09,716 cisTarget INFO Running cisTarget for topics_otsu_Topic21 which has 11230 regions
2024-03-11 15:17:14,426 cisTarget INFO Annotating motifs for topics_otsu_Topic31
2024-03-11 15:17:16,928 cisTarget INFO Getting cistromes for topics_otsu_Topic31
2024-03-11 15:17:17,245 cisTarget INFO Annotating motifs for topics_otsu_Topic2
2024-03-11 15:17:17,872 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:19,128 cisTarget INFO Getting cistromes for topics_otsu_Topic2
2024-03-11 15:17:19,611 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:20,633 cisTarget INFO Annotating motifs for topics_otsu_Topic12
2024-03-11 15:17:21,345 cisTarget INFO Annotating motifs for topics_otsu_Topic11
2024-03-11 15:17:21,878 cisTarget INFO Annotating motifs for topics_otsu_Topic35
2024-03-11 15:17:21,970 cisTarget INFO Annotating motifs for topics_otsu_Topic17
2024-03-11 15:17:22,530 cisTarget INFO Annotating motifs for topics_otsu_Topic37
2024-03-11 15:17:22,678 cisTarget INFO Annotating motifs for topics_otsu_Topic9
2024-03-11 15:17:22,902 cisTarget INFO Annotating motifs for topics_otsu_Topic25
2024-03-11 15:17:22,958 cisTarget INFO Annotating motifs for topics_otsu_Topic5
2024-03-11 15:17:23,288 cisTarget INFO Getting cistromes for topics_otsu_Topic12
2024-03-11 15:17:23,311 cisTarget INFO Annotating motifs for topics_otsu_Topic26
2024-03-11 15:17:23,327 cisTarget INFO Annotating motifs for topics_otsu_Topic33
2024-03-11 15:17:24,120 cisTarget INFO Getting cistromes for topics_otsu_Topic11
2024-03-11 15:17:24,137 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:24,363 cisTarget INFO Annotating motifs for topics_otsu_Topic30
2024-03-11 15:17:24,397 cisTarget INFO Getting cistromes for topics_otsu_Topic17
2024-03-11 15:17:24,491 cisTarget INFO Annotating motifs for topics_otsu_Topic28
2024-03-11 15:17:24,509 cisTarget INFO Annotating motifs for topics_otsu_Topic40
2024-03-11 15:17:24,570 cisTarget INFO Getting cistromes for topics_otsu_Topic35
2024-03-11 15:17:24,981 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:25,051 cisTarget INFO Getting cistromes for topics_otsu_Topic37
2024-03-11 15:17:25,114 cisTarget INFO Getting cistromes for topics_otsu_Topic9
2024-03-11 15:17:25,333 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:25,423 cisTarget INFO Getting cistromes for topics_otsu_Topic25
2024-03-11 15:17:25,597 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:25,624 cisTarget INFO Annotating motifs for topics_otsu_Topic15
2024-03-11 15:17:25,667 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:25,674 cisTarget INFO Getting cistromes for topics_otsu_Topic5
2024-03-11 15:17:25,698 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:25,701 cisTarget INFO Annotating motifs for topics_otsu_Topic29
2024-03-11 15:17:25,769 cisTarget INFO Getting cistromes for topics_otsu_Topic33
2024-03-11 15:17:25,947 cisTarget INFO Getting cistromes for topics_otsu_Topic26
2024-03-11 15:17:26,043 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:26,487 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:26,601 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:26,681 cisTarget INFO Getting cistromes for topics_otsu_Topic30
2024-03-11 15:17:26,775 cisTarget INFO Getting cistromes for topics_otsu_Topic40
2024-03-11 15:17:26,800 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:26,835 cisTarget INFO Getting cistromes for topics_otsu_Topic28
2024-03-11 15:17:26,855 cisTarget INFO Annotating motifs for topics_otsu_Topic38
2024-03-11 15:17:26,869 cisTarget INFO Running cisTarget for topics_otsu_Topic24 which has 3119 regions
2024-03-11 15:17:27,047 cisTarget INFO Annotating motifs for topics_otsu_Topic1
2024-03-11 15:17:27,134 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:27,231 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:27,378 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:27,490 cisTarget INFO Running cisTarget for topics_otsu_Topic20 which has 11197 regions
2024-03-11 15:17:27,816 cisTarget INFO Getting cistromes for topics_otsu_Topic15
2024-03-11 15:17:27,960 cisTarget INFO Getting cistromes for topics_otsu_Topic29
2024-03-11 15:17:28,268 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:28,464 cisTarget INFO Annotating motifs for topics_otsu_Topic21
2024-03-11 15:17:28,527 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:29,202 cisTarget INFO Getting cistromes for topics_otsu_Topic38
2024-03-11 15:17:29,405 cisTarget INFO Getting cistromes for topics_otsu_Topic1
2024-03-11 15:17:29,914 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:30,076 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:30,984 cisTarget INFO Getting cistromes for topics_otsu_Topic21
2024-03-11 15:17:31,814 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:32,001 cisTarget INFO Annotating motifs for topics_otsu_Topic24
2024-03-11 15:17:32,227 cisTarget INFO Running cisTarget for topics_otsu_Topic14 which has 2051 regions
2024-03-11 15:17:33,343 cisTarget INFO Running cisTarget for topics_otsu_Topic10 which has 1202 regions
2024-03-11 15:17:33,577 cisTarget INFO Running cisTarget for topics_otsu_Topic27 which has 3025 regions
2024-03-11 15:17:34,071 cisTarget INFO Getting cistromes for topics_otsu_Topic24
2024-03-11 15:17:34,177 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:34,315 cisTarget INFO Running cisTarget for topics_otsu_Topic8 which has 3054 regions
2024-03-11 15:17:34,340 cisTarget INFO Annotating motifs for topics_otsu_Topic20
2024-03-11 15:17:34,881 cisTarget INFO Running cisTarget for topics_otsu_Topic36 which has 5903 regions
2024-03-11 15:17:35,509 cisTarget INFO Running cisTarget for topics_otsu_Topic19 which has 6285 regions
2024-03-11 15:17:35,625 cisTarget INFO Running cisTarget for topics_otsu_Topic3 which has 3068 regions
2024-03-11 15:17:35,915 cisTarget INFO Running cisTarget for topics_otsu_Topic18 which has 4925 regions
2024-03-11 15:17:36,292 cisTarget INFO Getting cistromes for topics_otsu_Topic20
2024-03-11 15:17:36,347 cisTarget INFO Running cisTarget for topics_otsu_Topic22 which has 5026 regions
2024-03-11 15:17:36,553 cisTarget INFO Running cisTarget for topics_otsu_Topic7 which has 1260 regions
2024-03-11 15:17:36,555 cisTarget INFO Running cisTarget for topics_otsu_Topic39 which has 3328 regions
2024-03-11 15:17:36,818 cisTarget INFO Running cisTarget for topics_otsu_Topic4 which has 4995 regions
2024-03-11 15:17:36,958 cisTarget INFO Running cisTarget for topics_otsu_Topic16 which has 6702 regions
2024-03-11 15:17:37,176 cisTarget INFO Running cisTarget for topics_otsu_Topic23 which has 12440 regions
2024-03-11 15:17:37,728 cisTarget INFO Running cisTarget for topics_otsu_Topic32 which has 3980 regions
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/joblib/externals/loky/process_executor.py:752: UserWarning: A worker stopped while some jobs were given to the executor. This can be caused by a too short worker timeout or by a memory leak.
warnings.warn(
2024-03-11 15:17:40,951 cisTarget INFO Running cisTarget for topics_otsu_Topic34 which has 7682 regions
2024-03-11 15:17:41,096 cisTarget INFO Annotating motifs for topics_otsu_Topic14
2024-03-11 15:17:41,662 cisTarget INFO Running cisTarget for topics_otsu_Topic13 which has 2560 regions
2024-03-11 15:17:42,047 cisTarget INFO Running cisTarget for topics_top_3k_Topic21 which has 2987 regions
2024-03-11 15:17:42,872 cisTarget INFO Running cisTarget for topics_otsu_Topic6 which has 10743 regions
2024-03-11 15:17:43,159 cisTarget INFO Annotating motifs for topics_otsu_Topic10
2024-03-11 15:17:43,779 cisTarget INFO Getting cistromes for topics_otsu_Topic14
2024-03-11 15:17:44,251 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:44,497 cisTarget INFO Annotating motifs for topics_otsu_Topic8
2024-03-11 15:17:44,839 cisTarget INFO Annotating motifs for topics_otsu_Topic27
2024-03-11 15:17:45,830 cisTarget INFO Getting cistromes for topics_otsu_Topic10
2024-03-11 15:17:46,197 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:46,530 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:46,622 cisTarget INFO Annotating motifs for topics_otsu_Topic3
2024-03-11 15:17:46,979 cisTarget INFO Annotating motifs for topics_otsu_Topic18
2024-03-11 15:17:47,012 cisTarget INFO Annotating motifs for topics_otsu_Topic19
2024-03-11 15:17:47,044 cisTarget INFO Annotating motifs for topics_otsu_Topic39
2024-03-11 15:17:47,061 cisTarget INFO Getting cistromes for topics_otsu_Topic8
2024-03-11 15:17:47,157 cisTarget INFO Annotating motifs for topics_top_3k_Topic21
2024-03-11 15:17:47,210 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:47,414 cisTarget INFO Annotating motifs for topics_otsu_Topic36
2024-03-11 15:17:47,608 cisTarget INFO Annotating motifs for topics_otsu_Topic7
2024-03-11 15:17:48,009 cisTarget INFO Getting cistromes for topics_otsu_Topic27
2024-03-11 15:17:48,054 cisTarget INFO Annotating motifs for topics_otsu_Topic32
2024-03-11 15:17:48,124 cisTarget INFO Annotating motifs for topics_otsu_Topic22
2024-03-11 15:17:48,835 cisTarget INFO Annotating motifs for topics_otsu_Topic4
2024-03-11 15:17:48,891 cisTarget INFO Annotating motifs for topics_otsu_Topic16
2024-03-11 15:17:48,993 cisTarget INFO Getting cistromes for topics_top_3k_Topic21
2024-03-11 15:17:48,999 cisTarget INFO Getting cistromes for topics_otsu_Topic3
2024-03-11 15:17:49,236 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:49,346 cisTarget INFO Getting cistromes for topics_otsu_Topic18
2024-03-11 15:17:49,384 cisTarget INFO Getting cistromes for topics_otsu_Topic39
2024-03-11 15:17:49,408 cisTarget INFO Getting cistromes for topics_otsu_Topic19
2024-03-11 15:17:49,445 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:49,463 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:49,893 cisTarget INFO Getting cistromes for topics_otsu_Topic36
2024-03-11 15:17:50,159 cisTarget INFO Getting cistromes for topics_otsu_Topic7
2024-03-11 15:17:50,282 cisTarget INFO Getting cistromes for topics_otsu_Topic32
2024-03-11 15:17:50,489 cisTarget INFO Getting cistromes for topics_otsu_Topic22
2024-03-11 15:17:50,853 cisTarget INFO Annotating motifs for topics_otsu_Topic23
2024-03-11 15:17:50,912 cisTarget INFO Annotating motifs for topics_otsu_Topic13
2024-03-11 15:17:51,104 cisTarget INFO Annotating motifs for topics_otsu_Topic34
2024-03-11 15:17:51,140 cisTarget INFO Getting cistromes for topics_otsu_Topic16
2024-03-11 15:17:51,292 cisTarget INFO Getting cistromes for topics_otsu_Topic4
2024-03-11 15:17:52,332 cisTarget INFO Annotating motifs for topics_otsu_Topic6
2024-03-11 15:17:53,185 cisTarget INFO Getting cistromes for topics_otsu_Topic23
2024-03-11 15:17:53,258 cisTarget INFO Getting cistromes for topics_otsu_Topic34
2024-03-11 15:17:53,422 cisTarget INFO Running cisTarget for topics_top_3k_Topic28 which has 2990 regions
2024-03-11 15:17:53,469 cisTarget INFO Getting cistromes for topics_otsu_Topic13
2024-03-11 15:17:54,418 cisTarget INFO Getting cistromes for topics_otsu_Topic6
2024-03-11 15:17:54,538 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:54,558 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:54,614 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:54,791 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:54,847 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:54,902 cisTarget INFO Running cisTarget for topics_top_3k_Topic38 which has 2981 regions
2024-03-11 15:17:54,970 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:55,030 cisTarget INFO Running cisTarget for topics_top_3k_Topic15 which has 2988 regions
2024-03-11 15:17:55,788 cisTarget INFO Running cisTarget for topics_top_3k_Topic2 which has 2989 regions
2024-03-11 15:17:57,467 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:57,641 cisTarget INFO Running cisTarget for topics_top_3k_Topic37 which has 2929 regions
2024-03-11 15:17:57,741 cisTarget INFO Running cisTarget for topics_top_3k_Topic5 which has 2879 regions
2024-03-11 15:17:57,745 cisTarget INFO Running cisTarget for topics_top_3k_Topic11 which has 2967 regions
2024-03-11 15:17:58,942 cisTarget INFO Reading cisTarget database
2024-03-11 15:17:59,460 cisTarget INFO Annotating motifs for topics_top_3k_Topic28
2024-03-11 15:18:01,386 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:01,456 cisTarget INFO Annotating motifs for topics_top_3k_Topic15
2024-03-11 15:18:01,935 cisTarget INFO Getting cistromes for topics_top_3k_Topic28
2024-03-11 15:18:02,666 cisTarget INFO Annotating motifs for topics_top_3k_Topic2
2024-03-11 15:18:03,092 cisTarget INFO Running cisTarget for topics_top_3k_Topic12 which has 2954 regions
2024-03-11 15:18:03,593 cisTarget INFO Running cisTarget for topics_top_3k_Topic30 which has 2977 regions
2024-03-11 15:18:03,766 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:03,781 cisTarget INFO Running cisTarget for topics_top_3k_Topic1 which has 2891 regions
2024-03-11 15:18:03,919 cisTarget INFO Getting cistromes for topics_top_3k_Topic15
2024-03-11 15:18:04,097 cisTarget INFO Running cisTarget for topics_top_3k_Topic25 which has 2981 regions
2024-03-11 15:18:04,142 cisTarget INFO Running cisTarget for topics_top_3k_Topic26 which has 2978 regions
2024-03-11 15:18:04,480 cisTarget INFO Running cisTarget for topics_top_3k_Topic31 which has 2992 regions
2024-03-11 15:18:04,943 cisTarget INFO Annotating motifs for topics_top_3k_Topic37
2024-03-11 15:18:04,997 cisTarget INFO Annotating motifs for topics_top_3k_Topic11
2024-03-11 15:18:05,043 cisTarget INFO Getting cistromes for topics_top_3k_Topic2
2024-03-11 15:18:05,148 cisTarget INFO Annotating motifs for topics_top_3k_Topic38
2024-03-11 15:18:05,949 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:06,075 cisTarget INFO Annotating motifs for topics_top_3k_Topic5
2024-03-11 15:18:06,490 cisTarget INFO Running cisTarget for topics_top_3k_Topic40 which has 2993 regions
2024-03-11 15:18:07,319 cisTarget INFO Getting cistromes for topics_top_3k_Topic37
2024-03-11 15:18:07,383 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:07,490 cisTarget INFO Getting cistromes for topics_top_3k_Topic38
2024-03-11 15:18:07,520 cisTarget INFO Getting cistromes for topics_top_3k_Topic11
2024-03-11 15:18:08,037 cisTarget INFO Running cisTarget for topics_top_3k_Topic33 which has 2979 regions
2024-03-11 15:18:08,861 cisTarget INFO Getting cistromes for topics_top_3k_Topic5
2024-03-11 15:18:08,937 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:09,638 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:10,345 cisTarget INFO Annotating motifs for topics_top_3k_Topic1
2024-03-11 15:18:10,556 cisTarget INFO Running cisTarget for topics_top_3k_Topic35 which has 2973 regions
2024-03-11 15:18:10,767 cisTarget INFO Annotating motifs for topics_top_3k_Topic12
2024-03-11 15:18:11,301 cisTarget INFO Annotating motifs for topics_top_3k_Topic30
2024-03-11 15:18:11,633 cisTarget INFO Running cisTarget for topics_top_3k_Topic17 which has 2979 regions
2024-03-11 15:18:11,639 cisTarget INFO Annotating motifs for topics_top_3k_Topic25
2024-03-11 15:18:11,782 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:11,962 cisTarget INFO Annotating motifs for topics_top_3k_Topic26
2024-03-11 15:18:12,070 cisTarget INFO Annotating motifs for topics_top_3k_Topic31
2024-03-11 15:18:12,506 cisTarget INFO Getting cistromes for topics_top_3k_Topic1
2024-03-11 15:18:13,224 cisTarget INFO Getting cistromes for topics_top_3k_Topic12
2024-03-11 15:18:13,703 cisTarget INFO Getting cistromes for topics_top_3k_Topic30
2024-03-11 15:18:13,888 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:14,065 cisTarget INFO Getting cistromes for topics_top_3k_Topic25
2024-03-11 15:18:14,451 cisTarget INFO Getting cistromes for topics_top_3k_Topic26
2024-03-11 15:18:14,747 cisTarget INFO Getting cistromes for topics_top_3k_Topic31
2024-03-11 15:18:14,846 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:15,246 cisTarget INFO Running cisTarget for topics_top_3k_Topic9 which has 2886 regions
2024-03-11 15:18:15,497 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:16,144 cisTarget INFO Running cisTarget for topics_top_3k_Topic29 which has 2987 regions
2024-03-11 15:18:16,190 cisTarget INFO Annotating motifs for topics_top_3k_Topic40
2024-03-11 15:18:16,213 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:17,111 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:17,492 cisTarget INFO Running cisTarget for topics_top_3k_Topic20 which has 2988 regions
2024-03-11 15:18:17,705 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:18,149 cisTarget INFO Running cisTarget for topics_top_3k_Topic36 which has 2994 regions
2024-03-11 15:18:18,373 cisTarget INFO Annotating motifs for topics_top_3k_Topic33
2024-03-11 15:18:18,406 cisTarget INFO Getting cistromes for topics_top_3k_Topic40
2024-03-11 15:18:19,966 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:20,227 cisTarget INFO Annotating motifs for topics_top_3k_Topic35
2024-03-11 15:18:20,282 cisTarget INFO Running cisTarget for topics_top_3k_Topic24 which has 2944 regions
2024-03-11 15:18:20,648 cisTarget INFO Annotating motifs for topics_top_3k_Topic17
2024-03-11 15:18:20,824 cisTarget INFO Getting cistromes for topics_top_3k_Topic33
2024-03-11 15:18:21,520 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:21,747 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:21,762 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:22,483 cisTarget INFO Running cisTarget for topics_top_3k_Topic14 which has 2966 regions
2024-03-11 15:18:22,823 cisTarget INFO Getting cistromes for topics_top_3k_Topic35
2024-03-11 15:18:23,015 cisTarget INFO Getting cistromes for topics_top_3k_Topic17
2024-03-11 15:18:23,526 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:23,616 cisTarget INFO Running cisTarget for topics_top_3k_Topic8 which has 2971 regions
2024-03-11 15:18:23,753 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:23,767 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:24,260 cisTarget INFO Running cisTarget for topics_top_3k_Topic18 which has 2953 regions
2024-03-11 15:18:24,676 cisTarget INFO Annotating motifs for topics_top_3k_Topic36
2024-03-11 15:18:24,969 cisTarget INFO Running cisTarget for topics_top_3k_Topic27 which has 2990 regions
2024-03-11 15:18:25,382 cisTarget INFO Annotating motifs for topics_top_3k_Topic9
2024-03-11 15:18:25,792 cisTarget INFO Annotating motifs for topics_top_3k_Topic29
2024-03-11 15:18:26,294 cisTarget INFO Running cisTarget for topics_top_3k_Topic22 which has 2978 regions
2024-03-11 15:18:26,410 cisTarget INFO Running cisTarget for topics_top_3k_Topic23 which has 2988 regions
2024-03-11 15:18:26,426 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:27,043 cisTarget INFO Getting cistromes for topics_top_3k_Topic36
2024-03-11 15:18:27,632 cisTarget INFO Annotating motifs for topics_top_3k_Topic20
2024-03-11 15:18:27,834 cisTarget INFO Getting cistromes for topics_top_3k_Topic9
2024-03-11 15:18:28,154 cisTarget INFO Getting cistromes for topics_top_3k_Topic29
2024-03-11 15:18:28,282 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:29,028 cisTarget INFO Running cisTarget for topics_top_3k_Topic10 which has 2937 regions
2024-03-11 15:18:29,790 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:29,829 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:29,926 cisTarget INFO Getting cistromes for topics_top_3k_Topic20
2024-03-11 15:18:30,134 cisTarget INFO Annotating motifs for topics_top_3k_Topic8
2024-03-11 15:18:30,329 cisTarget INFO Annotating motifs for topics_top_3k_Topic24
2024-03-11 15:18:30,386 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:30,448 cisTarget INFO Running cisTarget for topics_top_3k_Topic32 which has 2968 regions
2024-03-11 15:18:30,827 cisTarget INFO Running cisTarget for topics_top_3k_Topic19 which has 2935 regions
2024-03-11 15:18:30,839 cisTarget INFO Annotating motifs for topics_top_3k_Topic18
2024-03-11 15:18:30,990 cisTarget INFO Running cisTarget for topics_top_3k_Topic39 which has 2966 regions
2024-03-11 15:18:30,998 cisTarget INFO Running cisTarget for topics_top_3k_Topic7 which has 2962 regions
2024-03-11 15:18:32,460 cisTarget INFO Getting cistromes for topics_top_3k_Topic8
2024-03-11 15:18:32,547 cisTarget INFO Running cisTarget for topics_top_3k_Topic34 which has 2984 regions
2024-03-11 15:18:32,566 cisTarget INFO Running cisTarget for topics_top_3k_Topic16 which has 2980 regions
2024-03-11 15:18:32,634 cisTarget INFO Getting cistromes for topics_top_3k_Topic24
2024-03-11 15:18:32,879 cisTarget INFO Annotating motifs for topics_top_3k_Topic22
2024-03-11 15:18:33,213 cisTarget INFO Annotating motifs for topics_top_3k_Topic14
2024-03-11 15:18:33,235 cisTarget INFO Getting cistromes for topics_top_3k_Topic18
2024-03-11 15:18:33,366 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:35,333 cisTarget INFO Getting cistromes for topics_top_3k_Topic22
2024-03-11 15:18:35,447 cisTarget INFO Running cisTarget for topics_top_3k_Topic3 which has 2942 regions
2024-03-11 15:18:35,672 cisTarget INFO Getting cistromes for topics_top_3k_Topic14
2024-03-11 15:18:36,310 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:36,635 cisTarget INFO Annotating motifs for topics_top_3k_Topic7
2024-03-11 15:18:36,737 cisTarget INFO Annotating motifs for topics_top_3k_Topic27
2024-03-11 15:18:36,835 cisTarget INFO Annotating motifs for topics_top_3k_Topic23
2024-03-11 15:18:37,192 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:37,705 cisTarget INFO Annotating motifs for topics_top_3k_Topic39
2024-03-11 15:18:37,852 cisTarget INFO Annotating motifs for topics_top_3k_Topic32
2024-03-11 15:18:38,054 cisTarget INFO Running cisTarget for topics_top_3k_Topic4 which has 2881 regions
2024-03-11 15:18:38,588 cisTarget INFO Running cisTarget for topics_top_3k_Topic13 which has 2952 regions
2024-03-11 15:18:38,981 cisTarget INFO Getting cistromes for topics_top_3k_Topic7
2024-03-11 15:18:39,194 cisTarget INFO Getting cistromes for topics_top_3k_Topic23
2024-03-11 15:18:39,482 cisTarget INFO Running cisTarget for topics_top_3k_Topic6 which has 2989 regions
2024-03-11 15:18:39,663 cisTarget INFO Getting cistromes for topics_top_3k_Topic27
2024-03-11 15:18:39,996 cisTarget INFO Getting cistromes for topics_top_3k_Topic39
2024-03-11 15:18:40,151 cisTarget INFO Getting cistromes for topics_top_3k_Topic32
2024-03-11 15:18:40,201 cisTarget INFO Annotating motifs for topics_top_3k_Topic10
2024-03-11 15:18:40,239 cisTarget INFO Annotating motifs for topics_top_3k_Topic34
2024-03-11 15:18:40,607 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:40,775 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:41,097 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:41,284 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:41,662 cisTarget INFO Annotating motifs for topics_top_3k_Topic19
2024-03-11 15:18:42,163 cisTarget INFO Running cisTarget for DARs_cell_type_INH_PVALB which has 17227 regions
2024-03-11 15:18:42,682 cisTarget INFO Getting cistromes for topics_top_3k_Topic10
2024-03-11 15:18:42,729 cisTarget INFO Annotating motifs for topics_top_3k_Topic16
2024-03-11 15:18:42,898 cisTarget INFO Getting cistromes for topics_top_3k_Topic34
2024-03-11 15:18:42,931 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:43,899 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:43,919 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:43,967 cisTarget INFO Getting cistromes for topics_top_3k_Topic19
2024-03-11 15:18:44,511 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:45,007 cisTarget INFO Getting cistromes for topics_top_3k_Topic16
2024-03-11 15:18:45,455 cisTarget INFO Running cisTarget for DARs_cell_type_INH_SNCG which has 17813 regions
2024-03-11 15:18:45,570 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:45,861 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:46,027 cisTarget INFO Annotating motifs for topics_top_3k_Topic6
2024-03-11 15:18:46,293 cisTarget INFO Annotating motifs for topics_top_3k_Topic3
2024-03-11 15:18:46,676 cisTarget INFO Annotating motifs for topics_top_3k_Topic13
2024-03-11 15:18:48,320 cisTarget INFO Getting cistromes for topics_top_3k_Topic6
2024-03-11 15:18:48,322 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:48,438 cisTarget INFO Running cisTarget for DARs_cell_type_MGL which has 9399 regions
2024-03-11 15:18:48,706 cisTarget INFO Getting cistromes for topics_top_3k_Topic3
2024-03-11 15:18:49,588 cisTarget INFO Getting cistromes for topics_top_3k_Topic13
2024-03-11 15:18:49,590 cisTarget INFO Running cisTarget for DARs_cell_type_MOL which has 25848 regions
2024-03-11 15:18:49,660 cisTarget INFO Annotating motifs for topics_top_3k_Topic4
2024-03-11 15:18:50,271 cisTarget INFO Reading cisTarget database
2024-03-11 15:18:52,147 cisTarget INFO Getting cistromes for topics_top_3k_Topic4
2024-03-11 15:18:52,732 cisTarget INFO No enriched motifs found for DARs_cell_type_MGL
2024-03-11 15:18:52,809 cisTarget INFO Annotating motifs for DARs_cell_type_INH_PVALB
2024-03-11 15:18:52,882 cisTarget INFO Running cisTarget for DARs_cell_type_GC which has 15356 regions
2024-03-11 15:18:53,190 cisTarget INFO Running cisTarget for DARs_cell_type_INH_VIP which has 18529 regions
2024-03-11 15:18:53,451 cisTarget INFO Running cisTarget for DARs_cell_type_COP which has 20350 regions
2024-03-11 15:18:53,841 cisTarget INFO Running cisTarget for DARs_cell_type_BG which has 30393 regions
2024-03-11 15:18:54,595 cisTarget INFO Running cisTarget for DARs_cell_type_OPC which has 11682 regions
2024-03-11 15:18:55,188 cisTarget INFO Getting cistromes for DARs_cell_type_INH_PVALB
2024-03-11 15:18:55,205 cisTarget INFO Running cisTarget for DARs_cell_type_INH_SST which has 17421 regions
2024-03-11 15:18:55,617 cisTarget INFO Running cisTarget for DARs_cell_type_PURK which has 19052 regions
2024-03-11 15:18:56,666 cisTarget INFO Running cisTarget for DARs_cell_type_NFOL which has 22425 regions
2024-03-11 15:18:57,096 cisTarget INFO Annotating motifs for DARs_cell_type_INH_SNCG
2024-03-11 15:18:57,326 cisTarget INFO Running cisTarget for DARs_cell_type_GP which has 16655 regions
2024-03-11 15:18:59,152 cisTarget INFO Running cisTarget for DARs_cell_type_AST which has 30080 regions
2024-03-11 15:18:59,401 cisTarget INFO Getting cistromes for DARs_cell_type_INH_SNCG
2024-03-11 15:19:01,654 cisTarget INFO Running cisTarget for DARs_cell_type_ENDO which has 14153 regions
2024-03-11 15:19:01,814 cisTarget INFO Running cisTarget for DARs_cell_type_MG which has 29511 regions
2024-03-11 15:19:05,149 cisTarget INFO Annotating motifs for DARs_cell_type_GC
2024-03-11 15:19:05,780 cisTarget INFO Annotating motifs for DARs_cell_type_OPC
2024-03-11 15:19:06,138 cisTarget INFO Annotating motifs for DARs_cell_type_INH_VIP
2024-03-11 15:19:06,162 cisTarget INFO No enriched motifs found for DARs_cell_type_ENDO
2024-03-11 15:19:07,282 cisTarget INFO Getting cistromes for DARs_cell_type_GC
2024-03-11 15:19:07,579 cisTarget INFO Annotating motifs for DARs_cell_type_MOL
2024-03-11 15:19:07,857 cisTarget INFO Getting cistromes for DARs_cell_type_OPC
2024-03-11 15:19:07,883 cisTarget INFO Annotating motifs for DARs_cell_type_BG
2024-03-11 15:19:08,262 cisTarget INFO Getting cistromes for DARs_cell_type_INH_VIP
2024-03-11 15:19:08,897 cisTarget INFO Annotating motifs for DARs_cell_type_COP
2024-03-11 15:19:09,024 cisTarget INFO Annotating motifs for DARs_cell_type_PURK
2024-03-11 15:19:09,144 cisTarget INFO Annotating motifs for DARs_cell_type_GP
2024-03-11 15:19:10,024 cisTarget INFO Getting cistromes for DARs_cell_type_MOL
2024-03-11 15:19:10,183 cisTarget INFO Getting cistromes for DARs_cell_type_BG
2024-03-11 15:19:10,876 cisTarget INFO Annotating motifs for DARs_cell_type_INH_SST
2024-03-11 15:19:11,064 cisTarget INFO Getting cistromes for DARs_cell_type_PURK
2024-03-11 15:19:11,126 cisTarget INFO Getting cistromes for DARs_cell_type_GP
2024-03-11 15:19:11,257 cisTarget INFO Getting cistromes for DARs_cell_type_COP
2024-03-11 15:19:12,058 cisTarget INFO Annotating motifs for DARs_cell_type_NFOL
2024-03-11 15:19:12,889 cisTarget INFO Getting cistromes for DARs_cell_type_INH_SST
2024-03-11 15:19:14,380 cisTarget INFO Getting cistromes for DARs_cell_type_NFOL
2024-03-11 15:19:16,336 cisTarget INFO Annotating motifs for DARs_cell_type_AST
2024-03-11 15:19:17,293 cisTarget INFO Annotating motifs for DARs_cell_type_MG
2024-03-11 15:19:18,369 cisTarget INFO Getting cistromes for DARs_cell_type_AST
2024-03-11 15:19:19,457 cisTarget INFO Getting cistromes for DARs_cell_type_MG
2024-03-11 15:19:20,871 SCENIC+ INFO Writing html to: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/ctx_results.html
2024-03-11 15:19:21,385 SCENIC+ INFO Writing output to: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/ctx_results.hdf5
[Mon Mar 11 15:19:42 2024]
Finished job 9.
1 of 13 steps (8%) done
Select jobs to execute...
Execute 1 jobs...
[Mon Mar 11 15:19:42 2024]
localrule download_genome_annotations:
output: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/genome_annotation.tsv, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/chromsizes.tsv
jobid: 8
reason: Params have changed since last execution
resources: tmpdir=/tmp
2024-03-11 15:20:03,500 Download gene annotation INFO Using genome: GRCh38.p14
2024-03-11 15:20:04,112 Download gene annotation INFO Found corresponding genome Id 51 on NCBI
2024-03-11 15:20:05,268 Download gene annotation INFO Found corresponding assembly Id 11968211 on NCBI
2024-03-11 15:20:06,251 Download gene annotation INFO Downloading assembly information from: ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/001/405/GCF_000001405.40_GRCh38.p14/GCF_000001405.40_GRCh38.p14_assembly_report.txt
2024-03-11 15:20:37,276 Download gene annotation INFO Found following assembled molecules (chromosomes):
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
X
Y
MT
2024-03-11 15:20:37,293 Download gene annotation INFO Converting chromosomes names to UCSC style as follows:
Original UCSC
1 chr1
2 chr2
3 chr3
4 chr4
5 chr5
6 chr6
7 chr7
8 chr8
9 chr9
10 chr10
11 chr11
12 chr12
13 chr13
14 chr14
15 chr15
16 chr16
17 chr17
18 chr18
19 chr19
20 chr20
21 chr21
22 chr22
X chrX
Y chrY
MT chrM
2024-03-11 15:20:37,311 SCENIC+ INFO Saving chromosome sizes to: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/chromsizes.tsv
2024-03-11 15:20:37,326 SCENIC+ INFO Saving genome annotation to: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/genome_annotation.tsv
[Mon Mar 11 15:20:38 2024]
Finished job 8.
2 of 13 steps (15%) done
Select jobs to execute...
Execute 1 jobs...
[Mon Mar 11 15:20:38 2024]
localrule motif_enrichment_dem:
input: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/data/region_sets, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/ctx_db/10x_brain_1kb_bg_with_mask.regions_vs_motifs.scores.feather, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/genome_annotation.tsv, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/ctx_db/aertslab_motif_colleciton/v10nr_clust_public/snapshots/motifs-v10-nr.hgnc-m0.00001-o0.0.tbl
output: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/dem_results.hdf5, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/dem_results.html
jobid: 7
reason: Input files updated by another job: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/genome_annotation.tsv
threads: 20
resources: tmpdir=/tmp
2024-03-11 15:20:43,090 SCENIC+ INFO Reading region sets from: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/data/region_sets
2024-03-11 15:20:43,091 SCENIC+ INFO Reading all .bed files in: topics_otsu
2024-03-11 15:20:43,318 SCENIC+ INFO Reading all .bed files in: topics_top_3k
2024-03-11 15:20:43,679 SCENIC+ INFO Reading all .bed files in: DARs_cell_type
2024-03-11 15:20:58,403 DEM INFO Running DEM for topics_otsu_Topic38_vs_all
2024-03-11 15:20:58,415 DEM INFO Running DEM for topics_otsu_Topic21_vs_all
2024-03-11 15:20:58,424 DEM INFO Running DEM for topics_otsu_Topic2_vs_all
2024-03-11 15:20:58,542 DEM INFO Running DEM for topics_otsu_Topic28_vs_all
2024-03-11 15:20:58,690 DEM INFO Running DEM for topics_otsu_Topic15_vs_all
2024-03-11 15:20:58,699 DEM INFO Running DEM for topics_otsu_Topic11_vs_all
2024-03-11 15:20:59,274 DEM INFO Running DEM for topics_otsu_Topic30_vs_all
2024-03-11 15:20:59,320 DEM INFO Running DEM for topics_otsu_Topic5_vs_all
2024-03-11 15:20:59,343 DEM INFO Running DEM for topics_otsu_Topic37_vs_all
2024-03-11 15:20:59,624 DEM INFO Running DEM for topics_otsu_Topic33_vs_all
2024-03-11 15:20:59,632 DEM INFO Running DEM for topics_otsu_Topic40_vs_all
2024-03-11 15:20:59,839 DEM INFO Running DEM for topics_otsu_Topic25_vs_all
2024-03-11 15:20:59,852 DEM INFO Running DEM for topics_otsu_Topic1_vs_all
2024-03-11 15:20:59,953 DEM INFO Running DEM for topics_otsu_Topic12_vs_all
2024-03-11 15:21:00,471 DEM INFO Running DEM for topics_otsu_Topic17_vs_all
2024-03-11 15:21:00,514 DEM INFO Running DEM for topics_otsu_Topic35_vs_all
2024-03-11 15:21:00,644 DEM INFO Running DEM for topics_otsu_Topic31_vs_all
2024-03-11 15:21:01,094 DEM INFO Running DEM for topics_otsu_Topic29_vs_all
2024-03-11 15:21:01,214 DEM INFO Running DEM for topics_otsu_Topic26_vs_all
2024-03-11 15:21:01,551 DEM INFO Running DEM for topics_otsu_Topic9_vs_all
2024-03-11 15:21:17,197 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:18,186 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:18,506 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:19,356 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:20,232 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:20,632 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:21,157 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:22,035 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:22,913 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:23,782 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:24,130 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:24,989 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:26,444 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:26,613 DEM INFO Running DEM for topics_otsu_Topic20_vs_all
2024-03-11 15:21:27,376 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:27,951 DEM INFO Running DEM for topics_otsu_Topic24_vs_all
2024-03-11 15:21:28,096 DEM INFO Running DEM for topics_otsu_Topic14_vs_all
2024-03-11 15:21:28,160 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:28,907 DEM INFO Running DEM for topics_otsu_Topic27_vs_all
2024-03-11 15:21:29,694 DEM INFO Running DEM for topics_otsu_Topic36_vs_all
2024-03-11 15:21:30,386 DEM INFO Running DEM for topics_otsu_Topic23_vs_all
2024-03-11 15:21:31,546 DEM INFO Running DEM for topics_otsu_Topic10_vs_all
2024-03-11 15:21:32,220 DEM INFO Running DEM for topics_otsu_Topic8_vs_all
2024-03-11 15:21:32,678 DEM INFO Running DEM for topics_otsu_Topic19_vs_all
2024-03-11 15:21:32,875 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:33,363 DEM INFO Running DEM for topics_otsu_Topic18_vs_all
2024-03-11 15:21:34,131 DEM INFO Running DEM for topics_otsu_Topic22_vs_all
2024-03-11 15:21:34,644 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:34,657 DEM INFO Running DEM for topics_otsu_Topic16_vs_all
2024-03-11 15:21:34,714 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:36,411 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:36,458 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:36,528 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:36,738 DEM INFO Running DEM for topics_otsu_Topic3_vs_all
2024-03-11 15:21:37,617 DEM INFO Running DEM for topics_otsu_Topic4_vs_all
2024-03-11 15:21:38,263 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:38,670 DEM INFO Running DEM for topics_otsu_Topic39_vs_all
2024-03-11 15:21:39,151 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:39,538 DEM INFO Adding motif-to-TF annotation
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/joblib/externals/loky/process_executor.py:752: UserWarning: A worker stopped while some jobs were given to the executor. This can be caused by a too short worker timeout or by a memory leak.
warnings.warn(
2024-03-11 15:21:42,171 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:42,747 DEM INFO Running DEM for topics_otsu_Topic32_vs_all
2024-03-11 15:21:44,368 DEM INFO Running DEM for topics_otsu_Topic7_vs_all
2024-03-11 15:21:44,377 DEM INFO Running DEM for topics_otsu_Topic34_vs_all
2024-03-11 15:21:46,264 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:46,297 DEM INFO Running DEM for topics_otsu_Topic6_vs_all
2024-03-11 15:21:46,346 DEM INFO Running DEM for topics_otsu_Topic13_vs_all
2024-03-11 15:21:46,813 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:47,866 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:48,170 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:49,008 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:49,121 DEM INFO Running DEM for topics_top_3k_Topic38_vs_all
2024-03-11 15:21:49,470 DEM INFO Running DEM for topics_top_3k_Topic15_vs_all
2024-03-11 15:21:49,624 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:50,997 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:51,158 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:51,617 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:53,032 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:54,580 DEM INFO Running DEM for topics_top_3k_Topic21_vs_all
2024-03-11 15:21:54,852 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:55,652 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:56,061 DEM INFO Running DEM for topics_top_3k_Topic11_vs_all
2024-03-11 15:21:56,426 DEM INFO Running DEM for topics_top_3k_Topic28_vs_all
2024-03-11 15:21:58,829 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:59,112 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:59,174 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:21:59,563 DEM INFO Running DEM for topics_top_3k_Topic5_vs_all
2024-03-11 15:22:00,244 DEM INFO Running DEM for topics_top_3k_Topic33_vs_all
2024-03-11 15:22:00,873 DEM INFO Running DEM for topics_top_3k_Topic2_vs_all
2024-03-11 15:22:02,913 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:04,302 DEM INFO Running DEM for topics_top_3k_Topic37_vs_all
2024-03-11 15:22:05,367 DEM INFO Running DEM for topics_top_3k_Topic35_vs_all
2024-03-11 15:22:05,688 DEM INFO Running DEM for topics_top_3k_Topic40_vs_all
2024-03-11 15:22:05,774 DEM INFO Running DEM for topics_top_3k_Topic26_vs_all
2024-03-11 15:22:05,957 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:07,578 DEM INFO Running DEM for topics_top_3k_Topic30_vs_all
2024-03-11 15:22:09,328 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:09,845 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:10,050 DEM INFO Running DEM for topics_top_3k_Topic1_vs_all
2024-03-11 15:22:10,258 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:10,902 DEM INFO Running DEM for topics_top_3k_Topic12_vs_all
2024-03-11 15:22:12,588 DEM INFO Running DEM for topics_top_3k_Topic25_vs_all
2024-03-11 15:22:13,093 DEM INFO Running DEM for topics_top_3k_Topic29_vs_all
2024-03-11 15:22:14,166 DEM INFO Running DEM for topics_top_3k_Topic31_vs_all
2024-03-11 15:22:14,589 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:15,285 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:15,914 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:16,469 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:17,077 DEM INFO Running DEM for topics_top_3k_Topic17_vs_all
2024-03-11 15:22:18,342 DEM INFO Running DEM for topics_top_3k_Topic9_vs_all
2024-03-11 15:22:19,710 DEM INFO Running DEM for topics_top_3k_Topic20_vs_all
2024-03-11 15:22:20,716 DEM INFO Running DEM for topics_top_3k_Topic14_vs_all
2024-03-11 15:22:21,146 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:23,103 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:23,970 DEM INFO Running DEM for topics_top_3k_Topic23_vs_all
2024-03-11 15:22:24,175 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:24,374 DEM INFO Running DEM for topics_top_3k_Topic24_vs_all
2024-03-11 15:22:25,707 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:26,424 DEM INFO Running DEM for topics_top_3k_Topic8_vs_all
2024-03-11 15:22:27,075 DEM INFO Running DEM for topics_top_3k_Topic10_vs_all
2024-03-11 15:22:27,568 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:27,802 DEM INFO Running DEM for topics_top_3k_Topic27_vs_all
2024-03-11 15:22:29,365 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:30,324 DEM INFO Running DEM for topics_top_3k_Topic18_vs_all
2024-03-11 15:22:30,874 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:30,884 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:31,021 DEM INFO Running DEM for topics_top_3k_Topic36_vs_all
2024-03-11 15:22:32,913 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:33,660 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:33,802 DEM INFO Running DEM for topics_top_3k_Topic19_vs_all
2024-03-11 15:22:34,313 DEM INFO Running DEM for topics_top_3k_Topic16_vs_all
2024-03-11 15:22:34,539 DEM INFO Running DEM for topics_top_3k_Topic3_vs_all
2024-03-11 15:22:34,647 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:36,385 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:36,711 DEM INFO Running DEM for topics_top_3k_Topic4_vs_all
2024-03-11 15:22:37,486 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:37,491 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:37,801 DEM INFO Running DEM for topics_top_3k_Topic39_vs_all
2024-03-11 15:22:38,656 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:39,985 DEM INFO Running DEM for topics_top_3k_Topic32_vs_all
2024-03-11 15:22:40,050 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:40,294 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:41,916 DEM INFO Running DEM for topics_top_3k_Topic22_vs_all
2024-03-11 15:22:42,574 DEM INFO Running DEM for topics_top_3k_Topic34_vs_all
2024-03-11 15:22:43,969 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:44,012 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:44,472 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:44,922 DEM INFO Running DEM for topics_top_3k_Topic13_vs_all
2024-03-11 15:22:46,424 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:46,945 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:47,587 DEM INFO Running DEM for DARs_cell_type_INH_PVALB_vs_all
2024-03-11 15:22:47,757 DEM INFO Running DEM for DARs_cell_type_INH_SNCG_vs_all
2024-03-11 15:22:48,333 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:49,387 DEM INFO Running DEM for topics_top_3k_Topic7_vs_all
2024-03-11 15:22:49,769 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:50,053 DEM INFO Running DEM for DARs_cell_type_MOL_vs_all
2024-03-11 15:22:50,212 DEM INFO Running DEM for DARs_cell_type_AST_vs_all
2024-03-11 15:22:51,755 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:51,777 DEM INFO Running DEM for topics_top_3k_Topic6_vs_all
2024-03-11 15:22:52,536 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:53,652 DEM INFO Running DEM for DARs_cell_type_BG_vs_all
2024-03-11 15:22:54,393 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:54,433 DEM INFO Running DEM for DARs_cell_type_MGL_vs_all
2024-03-11 15:22:55,076 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:22:57,090 DEM INFO Running DEM for DARs_cell_type_INH_SST_vs_all
2024-03-11 15:22:57,567 DEM INFO Running DEM for DARs_cell_type_GC_vs_all
2024-03-11 15:22:58,854 DEM INFO Running DEM for DARs_cell_type_ENDO_vs_all
2024-03-11 15:23:01,916 DEM INFO Running DEM for DARs_cell_type_PURK_vs_all
2024-03-11 15:23:02,891 DEM INFO Running DEM for DARs_cell_type_INH_VIP_vs_all
2024-03-11 15:23:04,004 DEM INFO Running DEM for DARs_cell_type_MG_vs_all
2024-03-11 15:23:04,202 DEM INFO Running DEM for DARs_cell_type_NFOL_vs_all
2024-03-11 15:23:04,381 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:23:06,266 DEM INFO Running DEM for DARs_cell_type_COP_vs_all
2024-03-11 15:23:09,092 DEM INFO Running DEM for DARs_cell_type_OPC_vs_all
2024-03-11 15:23:10,834 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:23:11,985 DEM INFO Running DEM for DARs_cell_type_GP_vs_all
2024-03-11 15:23:13,555 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:23:25,690 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:23:26,783 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:23:27,492 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:23:28,911 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:23:31,061 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:23:41,759 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:23:42,079 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:23:43,169 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:23:43,848 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:23:48,067 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:23:49,169 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:23:50,902 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:23:52,839 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:23:53,645 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:23:54,901 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:24:07,348 DEM INFO Adding motif-to-TF annotation
2024-03-11 15:24:09,105 SCENIC+ INFO Writing html to: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/dem_results.html
2024-03-11 15:24:09,152 SCENIC+ INFO Writing output to: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/dem_results.hdf5
[Mon Mar 11 15:24:12 2024]
Finished job 7.
3 of 13 steps (23%) done
Select jobs to execute...
Execute 2 jobs...
[Mon Mar 11 15:24:12 2024]
localrule prepare_menr:
input: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/dem_results.hdf5, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/ctx_results.hdf5, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/ACC_GEX.h5mu
output: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/tf_names.txt, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/cistromes_direct.h5ad, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/cistromes_extended.h5ad
jobid: 6
reason: Input files updated by another job: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/dem_results.hdf5, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/ctx_results.hdf5
resources: tmpdir=/tmp
[Mon Mar 11 15:24:12 2024]
localrule get_search_space:
input: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/ACC_GEX.h5mu, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/genome_annotation.tsv, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/chromsizes.tsv
output: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/search_space.tsv
jobid: 11
reason: Input files updated by another job: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/chromsizes.tsv, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/genome_annotation.tsv
resources: tmpdir=/tmp
2024-03-11 15:24:17,258 SCENIC+ INFO Reading multiome MuData.
2024-03-11 15:24:17,419 SCENIC+ INFO Reading data
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
2024-03-11 15:24:20,006 SCENIC+ INFO Getting cistromes.
2024-03-11 15:24:21,300 Get search space INFO Extending promoter annotation to 10 bp upstream and 10 downstream
2024-03-11 15:24:21,506 Get search space INFO Extending search space to:
150000 bp downstream of the end of the gene.
150000 bp upstream of the start of the gene.
2024-03-11 15:24:22,583 Get search space INFO Intersecting with regions.
2024-03-11 15:24:25,330 Get search space INFO Calculating distances from region to gene
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
2024-03-11 15:24:41,738 SCENIC+ INFO Found 788 TFs.
2024-03-11 15:24:41,738 SCENIC+ INFO Saving TF names to: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/tf_names.txt
2024-03-11 15:24:41,740 SCENIC+ INFO Writing direct cistromes to: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/cistromes_direct.h5ad
... storing 'motifs' as categorical
2024-03-11 15:24:41,897 SCENIC+ INFO Writing extended cistromes to: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/cistromes_extended.h5ad
... storing 'motifs' as categorical
[Mon Mar 11 15:24:42 2024]
Finished job 6.
4 of 13 steps (31%) done
Select jobs to execute...
2024-03-11 15:26:03,153 Get search space INFO Imploding multiple entries per region and gene
2024-03-11 15:29:00,177 SCENIC+ INFO Writing search space to: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/search_space.tsv
[Mon Mar 11 15:29:04 2024]
Finished job 11.
5 of 13 steps (38%) done
Execute 1 jobs...
[Mon Mar 11 15:29:04 2024]
localrule region_to_gene:
input: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/ACC_GEX.h5mu, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/search_space.tsv
output: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/region_to_gene_adj.tsv
jobid: 10
reason: Input files updated by another job: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/search_space.tsv
threads: 20
resources: tmpdir=/tmp
2024-03-11 15:29:09,335 SCENIC+ INFO Reading multiome MuData.
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
2024-03-11 15:29:11,956 SCENIC+ INFO Reading search space
2024-03-11 15:29:12,941 R2G INFO Calculating region to gene importances, using GBM method
Running using 20 cores: 100%|█████████████| 18569/18569 [07:26<00:00, 41.62it/s]
2024-03-11 15:36:48,829 R2G INFO Calculating region to gene correlation, using SR method
Running using 20 cores: 0%| | 0/18569 [00:00<?, ?it/s]/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/scipy/stats/_stats_py.py:4916: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
warnings.warn(stats.ConstantInputWarning(warn_msg))
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/scipy/stats/_stats_py.py:4916: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
warnings.warn(stats.ConstantInputWarning(warn_msg))
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/scipy/stats/_stats_py.py:4916: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
warnings.warn(stats.ConstantInputWarning(warn_msg))
Running using 20 cores: 1%| | 141/18569 [00:00<00:42, 429.72it/s]/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/scipy/stats/_stats_py.py:4916: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
warnings.warn(stats.ConstantInputWarning(warn_msg))
Running using 20 cores: 1%|▏ | 200/18569 [00:00<00:44, 408.27it/s]/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/scipy/stats/_stats_py.py:4916: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
warnings.warn(stats.ConstantInputWarning(warn_msg))
Running using 20 cores: 2%|▏ | 280/18569 [00:00<00:52, 350.98it/s]/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/scipy/stats/_stats_py.py:4916: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
warnings.warn(stats.ConstantInputWarning(warn_msg))
Running using 20 cores: 2%|▏ | 324/18569 [00:00<00:49, 369.51it/s]/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/scipy/stats/_stats_py.py:4916: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
warnings.warn(stats.ConstantInputWarning(warn_msg))
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/scipy/stats/_stats_py.py:4916: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
warnings.warn(stats.ConstantInputWarning(warn_msg))
Running using 20 cores: 2%|▎ | 364/18569 [00:01<00:56, 323.06it/s]/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/scipy/stats/_stats_py.py:4916: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
warnings.warn(stats.ConstantInputWarning(warn_msg))
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/scipy/stats/_stats_py.py:4916: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
warnings.warn(stats.ConstantInputWarning(warn_msg))
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/scipy/stats/_stats_py.py:4916: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
warnings.warn(stats.ConstantInputWarning(warn_msg))
Running using 20 cores: 2%|▎ | 440/18569 [00:01<00:57, 312.58it/s]/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/scipy/stats/_stats_py.py:4916: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
warnings.warn(stats.ConstantInputWarning(warn_msg))
Running using 20 cores: 3%|▍ | 596/18569 [00:01<00:53, 336.21it/s]/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/scipy/stats/_stats_py.py:4916: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
warnings.warn(stats.ConstantInputWarning(warn_msg))
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/scipy/stats/_stats_py.py:4916: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
warnings.warn(stats.ConstantInputWarning(warn_msg))
Running using 20 cores: 3%|▍ | 640/18569 [00:02<01:07, 264.12it/s]/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/scipy/stats/_stats_py.py:4916: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
warnings.warn(stats.ConstantInputWarning(warn_msg))
Running using 20 cores: 4%|▌ | 680/18569 [00:02<01:12, 247.59it/s]/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/scipy/stats/_stats_py.py:4916: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
warnings.warn(stats.ConstantInputWarning(warn_msg))
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/scipy/stats/_stats_py.py:4916: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
warnings.warn(stats.ConstantInputWarning(warn_msg))
Running using 20 cores: 4%|▌ | 760/18569 [00:02<00:59, 297.62it/s]/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/scipy/stats/_stats_py.py:4916: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
warnings.warn(stats.ConstantInputWarning(warn_msg))
Running using 20 cores: 6%|▊ | 1138/18569 [00:03<01:00, 289.83it/s]/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/scipy/stats/_stats_py.py:4916: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
warnings.warn(stats.ConstantInputWarning(warn_msg))
Running using 20 cores: 13%|█▋ | 2360/18569 [00:08<00:55, 292.27it/s]/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/scipy/stats/_stats_py.py:4916: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
warnings.warn(stats.ConstantInputWarning(warn_msg))
Running using 20 cores: 100%|████████████| 18569/18569 [01:02<00:00, 296.97it/s]
2024-03-11 15:38:12,496 R2G INFO Done!
2024-03-11 15:38:12,767 SCENIC+ INFO Saving region to gene adjacencies to /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/region_to_gene_adj.tsv
[Mon Mar 11 15:38:23 2024]
Finished job 10.
6 of 13 steps (46%) done
Select jobs to execute...
Execute 1 jobs...
[Mon Mar 11 15:38:23 2024]
localrule tf_to_gene:
input: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/ACC_GEX.h5mu, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/tf_names.txt
output: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/tf_to_gene_adj.tsv
jobid: 5
reason: Missing output files: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/tf_to_gene_adj.tsv; Input files updated by another job: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/tf_names.txt
threads: 20
resources: tmpdir=/tmp
2024-03-11 15:38:28,846 SCENIC+ INFO Reading multiome MuData.
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
2024-03-11 15:38:31,412 SCENIC+ INFO Using 789 TFs.
2024-03-11 15:38:31,809 TF2G INFO Calculating TF-to-gene importance
Running using 20 cores: 100%|█████████████| 36601/36601 [22:12<00:00, 27.47it/s]
2024-03-11 16:00:52,659 TF2G INFO Adding correlation coefficients to adjacencies.
2024-03-11 16:01:16,586 TF2G INFO Warning: adding TFs as their own target to adjecencies matrix. Importance values will be max + 1e-05
2024-03-11 16:01:23,858 SCENIC+ INFO Saving TF to gene adjacencies to: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/tf_to_gene_adj.tsv
[Mon Mar 11 16:01:33 2024]
Finished job 5.
7 of 13 steps (54%) done
Select jobs to execute...
Execute 1 jobs...
[Mon Mar 11 16:01:33 2024]
localrule eGRN_direct:
input: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/tf_to_gene_adj.tsv, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/region_to_gene_adj.tsv, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/cistromes_direct.h5ad, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/ctx_db/10x_brain_1kb_bg_with_mask.regions_vs_motifs.rankings.feather
output: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/eRegulon_direct.tsv
jobid: 4
reason: Missing output files: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/eRegulon_direct.tsv; Input files updated by another job: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/cistromes_direct.h5ad, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/region_to_gene_adj.tsv, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/tf_to_gene_adj.tsv
threads: 20
resources: tmpdir=/tmp
2024-03-11 16:01:39,749 SCENIC+ INFO Loading TF to gene adjacencies.
2024-03-11 16:01:41,133 SCENIC+ INFO Loading region to gene adjacencies.
2024-03-11 16:01:43,266 SCENIC+ INFO Loading cistromes.
2024-03-11 16:01:43,342 GSEA INFO Thresholding region to gene relationships
0%| | 0/14 [00:00<?, ?it/s]
Processing: negative r2g, 0.85 quantile: 0%| | 0/574 [00:00<?, ?it/s]
Processing: negative r2g, 0.85 quantile: 1%| | 8/574 [00:00<00:07, 72.03it/s]
Processing: negative r2g, 0.85 quantile: 3%| | 18/574 [00:00<00:06, 84.00it/s]
Processing: negative r2g, 0.85 quantile: 5%| | 29/574 [00:00<00:05, 93.15it/s]
Processing: negative r2g, 0.85 quantile: 7%| | 39/574 [00:00<00:06, 83.96it/s]
Processing: negative r2g, 0.85 quantile: 8%| | 48/574 [00:00<00:06, 82.57it/s]
Processing: negative r2g, 0.85 quantile: 10%| | 57/574 [00:00<00:06, 82.23it/s]
Processing: negative r2g, 0.85 quantile: 11%| | 66/574 [00:00<00:09, 51.84it/s]
Processing: negative r2g, 0.85 quantile: 13%|▏| 74/574 [00:01<00:08, 56.84it/s]
Processing: negative r2g, 0.85 quantile: 14%|▏| 83/574 [00:01<00:07, 63.05it/s]
Processing: negative r2g, 0.85 quantile: 16%|▏| 93/574 [00:01<00:06, 71.82it/s]
Processing: negative r2g, 0.85 quantile: 18%|▏| 102/574 [00:01<00:06, 75.35it/s
Processing: negative r2g, 0.85 quantile: 19%|▏| 111/574 [00:01<00:06, 72.00it/s
Processing: negative r2g, 0.85 quantile: 21%|▏| 119/574 [00:01<00:06, 72.61it/s
Processing: negative r2g, 0.85 quantile: 23%|▏| 132/574 [00:01<00:05, 82.71it/s
Processing: negative r2g, 0.85 quantile: 25%|▏| 141/574 [00:01<00:05, 83.23it/s
Processing: negative r2g, 0.85 quantile: 26%|▎| 150/574 [00:02<00:05, 82.73it/s
Processing: negative r2g, 0.85 quantile: 28%|▎| 159/574 [00:02<00:04, 83.30it/s
Processing: negative r2g, 0.85 quantile: 29%|▎| 168/574 [00:02<00:05, 74.27it/s
Processing: negative r2g, 0.85 quantile: 31%|▎| 178/574 [00:02<00:04, 80.60it/s
Processing: negative r2g, 0.85 quantile: 33%|▎| 187/574 [00:02<00:07, 54.44it/s
Processing: negative r2g, 0.85 quantile: 34%|▎| 195/574 [00:02<00:06, 58.61it/s
Processing: negative r2g, 0.85 quantile: 35%|▎| 203/574 [00:02<00:05, 62.84it/s
Processing: negative r2g, 0.85 quantile: 37%|▎| 211/574 [00:02<00:05, 64.00it/s
Processing: negative r2g, 0.85 quantile: 39%|▍| 221/574 [00:03<00:04, 71.12it/s
Processing: negative r2g, 0.85 quantile: 40%|▍| 229/574 [00:03<00:04, 72.57it/s
Processing: negative r2g, 0.85 quantile: 42%|▍| 239/574 [00:03<00:04, 78.64it/s
Processing: negative r2g, 0.85 quantile: 43%|▍| 249/574 [00:03<00:03, 83.29it/s
Processing: negative r2g, 0.85 quantile: 45%|▍| 258/574 [00:03<00:03, 82.38it/s
Processing: negative r2g, 0.85 quantile: 47%|▍| 267/574 [00:03<00:03, 78.41it/s
Processing: negative r2g, 0.85 quantile: 48%|▍| 276/574 [00:03<00:03, 78.90it/s
Processing: negative r2g, 0.85 quantile: 50%|▍| 285/574 [00:03<00:03, 81.43it/s
Processing: negative r2g, 0.85 quantile: 51%|▌| 295/574 [00:03<00:03, 86.14it/s
Processing: negative r2g, 0.85 quantile: 53%|▌| 304/574 [00:04<00:03, 85.86it/s
Processing: negative r2g, 0.85 quantile: 55%|▌| 313/574 [00:04<00:05, 52.17it/s
Processing: negative r2g, 0.85 quantile: 56%|▌| 320/574 [00:04<00:04, 52.65it/s
Processing: negative r2g, 0.85 quantile: 58%|▌| 331/574 [00:04<00:03, 62.01it/s
Processing: negative r2g, 0.85 quantile: 59%|▌| 341/574 [00:04<00:03, 69.88it/s
Processing: negative r2g, 0.85 quantile: 61%|▌| 351/574 [00:04<00:02, 76.53it/s
Processing: negative r2g, 0.85 quantile: 63%|▋| 360/574 [00:05<00:03, 68.02it/s
Processing: negative r2g, 0.85 quantile: 64%|▋| 368/574 [00:05<00:03, 68.37it/s
Processing: negative r2g, 0.85 quantile: 66%|▋| 377/574 [00:05<00:02, 70.77it/s
Processing: negative r2g, 0.85 quantile: 67%|▋| 385/574 [00:05<00:03, 62.66it/s
Processing: negative r2g, 0.85 quantile: 68%|▋| 392/574 [00:05<00:02, 63.13it/s
Processing: negative r2g, 0.85 quantile: 70%|▋| 400/574 [00:05<00:02, 66.66it/s
Processing: negative r2g, 0.85 quantile: 72%|▋| 411/574 [00:05<00:02, 75.91it/s
Processing: negative r2g, 0.85 quantile: 73%|▋| 420/574 [00:05<00:01, 79.24it/s
Processing: negative r2g, 0.85 quantile: 75%|▋| 429/574 [00:05<00:01, 81.67it/s
Processing: negative r2g, 0.85 quantile: 76%|▊| 439/574 [00:06<00:01, 85.36it/s
Processing: negative r2g, 0.85 quantile: 78%|▊| 448/574 [00:06<00:02, 47.97it/s
Processing: negative r2g, 0.85 quantile: 80%|▊| 458/574 [00:06<00:02, 56.77it/s
Processing: negative r2g, 0.85 quantile: 82%|▊| 469/574 [00:06<00:01, 67.18it/s
Processing: negative r2g, 0.85 quantile: 84%|▊| 480/574 [00:06<00:01, 75.70it/s
Processing: negative r2g, 0.85 quantile: 85%|▊| 490/574 [00:06<00:01, 72.63it/s
Processing: negative r2g, 0.85 quantile: 87%|▊| 500/574 [00:07<00:00, 77.59it/s
Processing: negative r2g, 0.85 quantile: 89%|▉| 510/574 [00:07<00:00, 81.51it/s
Processing: negative r2g, 0.85 quantile: 91%|▉| 521/574 [00:07<00:00, 88.46it/s
Processing: negative r2g, 0.85 quantile: 93%|▉| 531/574 [00:07<00:00, 90.96it/s
Processing: negative r2g, 0.85 quantile: 94%|▉| 541/574 [00:07<00:00, 80.98it/s
Processing: negative r2g, 0.85 quantile: 96%|▉| 553/574 [00:07<00:00, 89.10it/s
Processing: negative r2g, 0.85 quantile: 98%|▉| 564/574 [00:07<00:00, 93.60it/s
Processing: negative r2g, 0.85 quantile: 100%|█| 574/574 [00:07<00:00, 91.77it/s
7%|███▏ | 1/14 [00:10<02:12, 10.18s/it]
Processing: negative r2g, 0.9 quantile: 0%| | 0/574 [00:00<?, ?it/s]
Processing: negative r2g, 0.9 quantile: 2%| | 13/574 [00:00<00:04, 121.80it/s]
Processing: negative r2g, 0.9 quantile: 5%| | 27/574 [00:00<00:04, 129.28it/s]
Processing: negative r2g, 0.9 quantile: 7%| | 40/574 [00:00<00:04, 114.69it/s]
Processing: negative r2g, 0.9 quantile: 9%| | 52/574 [00:00<00:04, 115.63it/s]
Processing: negative r2g, 0.9 quantile: 11%| | 64/574 [00:00<00:04, 112.64it/s]
Processing: negative r2g, 0.9 quantile: 14%|▏| 78/574 [00:00<00:04, 118.37it/s]
Processing: negative r2g, 0.9 quantile: 16%|▏| 91/574 [00:00<00:03, 121.81it/s]
Processing: negative r2g, 0.9 quantile: 18%|▏| 104/574 [00:00<00:03, 118.23it/s
Processing: negative r2g, 0.9 quantile: 20%|▏| 116/574 [00:00<00:03, 115.42it/s
Processing: negative r2g, 0.9 quantile: 23%|▏| 132/574 [00:01<00:03, 122.54it/s
Processing: negative r2g, 0.9 quantile: 25%|▎| 145/574 [00:01<00:03, 120.15it/s
Processing: negative r2g, 0.9 quantile: 28%|▎| 158/574 [00:01<00:03, 119.61it/s
Processing: negative r2g, 0.9 quantile: 30%|▎| 170/574 [00:01<00:06, 59.66it/s]
Processing: negative r2g, 0.9 quantile: 32%|▎| 184/574 [00:01<00:05, 72.23it/s]
Processing: negative r2g, 0.9 quantile: 34%|▎| 195/574 [00:02<00:04, 77.94it/s]
Processing: negative r2g, 0.9 quantile: 36%|▎| 206/574 [00:02<00:04, 80.41it/s]
Processing: negative r2g, 0.9 quantile: 38%|▍| 220/574 [00:02<00:03, 92.11it/s]
Processing: negative r2g, 0.9 quantile: 41%|▍| 234/574 [00:02<00:03, 102.63it/s
Processing: negative r2g, 0.9 quantile: 43%|▍| 247/574 [00:02<00:03, 108.04it/s
Processing: negative r2g, 0.9 quantile: 45%|▍| 259/574 [00:02<00:02, 107.82it/s
Processing: negative r2g, 0.9 quantile: 48%|▍| 273/574 [00:02<00:02, 115.01it/s
Processing: negative r2g, 0.9 quantile: 50%|▍| 286/574 [00:02<00:02, 117.09it/s
Processing: negative r2g, 0.9 quantile: 52%|▌| 299/574 [00:02<00:02, 116.19it/s
Processing: negative r2g, 0.9 quantile: 54%|▌| 311/574 [00:02<00:02, 114.80it/s
Processing: negative r2g, 0.9 quantile: 56%|▌| 323/574 [00:03<00:02, 101.09it/s
Processing: negative r2g, 0.9 quantile: 58%|▌| 335/574 [00:03<00:02, 104.73it/s
Processing: negative r2g, 0.9 quantile: 61%|▌| 349/574 [00:03<00:01, 113.05it/s
Processing: negative r2g, 0.9 quantile: 63%|▋| 361/574 [00:03<00:01, 113.46it/s
Processing: negative r2g, 0.9 quantile: 65%|▋| 373/574 [00:03<00:01, 103.62it/s
Processing: negative r2g, 0.9 quantile: 67%|▋| 384/574 [00:03<00:01, 100.40it/s
Processing: negative r2g, 0.9 quantile: 69%|▋| 395/574 [00:03<00:01, 100.04it/s
Processing: negative r2g, 0.9 quantile: 71%|▋| 407/574 [00:03<00:01, 103.78it/s
Processing: negative r2g, 0.9 quantile: 73%|▋| 418/574 [00:04<00:01, 105.01it/s
Processing: negative r2g, 0.9 quantile: 75%|▊| 432/574 [00:04<00:01, 113.70it/s
Processing: negative r2g, 0.9 quantile: 77%|▊| 444/574 [00:04<00:01, 109.67it/s
Processing: negative r2g, 0.9 quantile: 79%|▊| 456/574 [00:04<00:01, 59.13it/s]
Processing: negative r2g, 0.9 quantile: 82%|▊| 470/574 [00:04<00:01, 72.47it/s]
Processing: negative r2g, 0.9 quantile: 84%|▊| 483/574 [00:04<00:01, 82.90it/s]
Processing: negative r2g, 0.9 quantile: 87%|▊| 497/574 [00:05<00:00, 92.75it/s]
Processing: negative r2g, 0.9 quantile: 89%|▉| 509/574 [00:05<00:00, 97.46it/s]
Processing: negative r2g, 0.9 quantile: 91%|▉| 522/574 [00:05<00:00, 104.74it/s
Processing: negative r2g, 0.9 quantile: 93%|▉| 535/574 [00:05<00:00, 109.27it/s
Processing: negative r2g, 0.9 quantile: 96%|▉| 550/574 [00:05<00:00, 118.15it/s
Processing: negative r2g, 0.9 quantile: 98%|▉| 564/574 [00:05<00:00, 122.43it/s
14%|██████▎ | 2/14 [00:16<01:35, 7.95s/it]
Processing: 0.95 quantile, negative r2g: 0%| | 0/574 [00:00<?, ?it/s]
Processing: 0.95 quantile, negative r2g: 3%| | 18/574 [00:00<00:03, 173.76it/s
Processing: 0.95 quantile, negative r2g: 6%| | 37/574 [00:00<00:02, 182.05it/s
Processing: 0.95 quantile, negative r2g: 10%| | 56/574 [00:00<00:02, 174.91it/s
Processing: 0.95 quantile, negative r2g: 13%|▏| 75/574 [00:00<00:02, 177.33it/s
Processing: 0.95 quantile, negative r2g: 17%|▏| 95/574 [00:00<00:02, 182.04it/s
Processing: 0.95 quantile, negative r2g: 20%|▏| 114/574 [00:00<00:02, 174.52it/
Processing: 0.95 quantile, negative r2g: 23%|▏| 134/574 [00:00<00:02, 180.33it/
Processing: 0.95 quantile, negative r2g: 27%|▎| 154/574 [00:00<00:02, 183.60it/
Processing: 0.95 quantile, negative r2g: 30%|▎| 173/574 [00:00<00:02, 175.78it/
Processing: 0.95 quantile, negative r2g: 33%|▎| 191/574 [00:01<00:02, 174.31it/
Processing: 0.95 quantile, negative r2g: 36%|▎| 209/574 [00:01<00:02, 171.82it/
Processing: 0.95 quantile, negative r2g: 40%|▍| 229/574 [00:01<00:01, 177.44it/
Processing: 0.95 quantile, negative r2g: 43%|▍| 249/574 [00:01<00:01, 180.66it/
Processing: 0.95 quantile, negative r2g: 47%|▍| 268/574 [00:01<00:01, 177.55it/
Processing: 0.95 quantile, negative r2g: 50%|▍| 286/574 [00:01<00:01, 176.87it/
Processing: 0.95 quantile, negative r2g: 53%|▌| 304/574 [00:01<00:01, 176.40it/
Processing: 0.95 quantile, negative r2g: 56%|▌| 322/574 [00:01<00:01, 165.40it/
Processing: 0.95 quantile, negative r2g: 59%|▌| 340/574 [00:01<00:01, 167.98it/
Processing: 0.95 quantile, negative r2g: 63%|▋| 359/574 [00:02<00:01, 172.59it/
Processing: 0.95 quantile, negative r2g: 66%|▋| 377/574 [00:02<00:01, 167.61it/
Processing: 0.95 quantile, negative r2g: 69%|▋| 394/574 [00:02<00:01, 158.63it/
Processing: 0.95 quantile, negative r2g: 72%|▋| 413/574 [00:02<00:00, 166.45it/
Processing: 0.95 quantile, negative r2g: 75%|▊| 433/574 [00:02<00:00, 173.87it/
Processing: 0.95 quantile, negative r2g: 79%|▊| 451/574 [00:02<00:00, 168.14it/
Processing: 0.95 quantile, negative r2g: 82%|▊| 472/574 [00:02<00:00, 179.24it/
Processing: 0.95 quantile, negative r2g: 86%|▊| 491/574 [00:03<00:00, 86.08it/s
Processing: 0.95 quantile, negative r2g: 89%|▉| 508/574 [00:03<00:00, 98.78it/s
Processing: 0.95 quantile, negative r2g: 92%|▉| 529/574 [00:03<00:00, 118.90it/
Processing: 0.95 quantile, negative r2g: 95%|▉| 548/574 [00:03<00:00, 133.53it/
Processing: 0.95 quantile, negative r2g: 99%|▉| 569/574 [00:03<00:00, 150.09it/
21%|█████████▍ | 3/14 [00:20<01:09, 6.32s/it]
Processing: negative r2g, Top 5 region-to-gene links per gene: 0%| | 0/574 [00
Processing: negative r2g, Top 5 region-to-gene links per gene: 1%| | 6/574 [00
Processing: negative r2g, Top 5 region-to-gene links per gene: 2%| | 14/574 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 4%| | 22/574 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 5%| | 30/574 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 7%| | 38/574 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 8%| | 46/574 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 9%| | 53/574 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 10%| | 60/574 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 12%| | 67/574 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 13%|▏| 75/574 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 14%|▏| 83/574 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 16%|▏| 91/574 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 17%|▏| 98/574 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 18%|▏| 105/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 20%|▏| 113/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 21%|▏| 121/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 22%|▏| 128/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 24%|▏| 135/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 25%|▏| 143/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 26%|▎| 150/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 28%|▎| 158/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 29%|▎| 165/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 30%|▎| 171/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 31%|▎| 179/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 32%|▎| 186/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 34%|▎| 193/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 35%|▎| 200/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 36%|▎| 207/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 37%|▎| 215/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 39%|▍| 222/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 40%|▍| 230/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 41%|▍| 238/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 43%|▍| 245/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 44%|▍| 252/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 45%|▍| 259/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 46%|▍| 266/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 47%|▍| 272/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 49%|▍| 280/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 50%|▌| 287/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 51%|▌| 294/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 52%|▌| 301/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 53%|▌| 307/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 55%|▌| 313/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 55%|▌| 318/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 57%|▌| 325/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 58%|▌| 331/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 59%|▌| 339/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 61%|▌| 348/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 62%|▌| 355/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 63%|▋| 362/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 64%|▋| 369/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 65%|▋| 375/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 66%|▋| 381/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 67%|▋| 386/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 68%|▋| 392/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 69%|▋| 397/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 71%|▋| 405/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 72%|▋| 413/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 73%|▋| 420/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 74%|▋| 427/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 76%|▊| 434/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 77%|▊| 441/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 78%|▊| 448/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 79%|▊| 455/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 80%|▊| 462/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 82%|▊| 471/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 83%|▊| 478/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 84%|▊| 485/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 86%|▊| 493/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 87%|▊| 501/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 89%|▉| 508/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 90%|▉| 515/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 91%|▉| 523/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 93%|▉| 532/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 94%|▉| 540/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 95%|▉| 547/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 97%|▉| 555/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 98%|▉| 564/574 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 99%|▉| 571/574 [
29%|████████████▌ | 4/14 [00:31<01:21, 8.11s/it]
Processing: negative r2g, Top 10 region-to-gene links per gene: 0%| | 0/574 [0
Processing: negative r2g, Top 10 region-to-gene links per gene: 1%| | 4/574 [0
Processing: negative r2g, Top 10 region-to-gene links per gene: 1%| | 8/574 [0
Processing: negative r2g, Top 10 region-to-gene links per gene: 2%| | 13/574 [
Processing: negative r2g, Top 10 region-to-gene links per gene: 3%| | 17/574 [
Processing: negative r2g, Top 10 region-to-gene links per gene: 4%| | 21/574 [
Processing: negative r2g, Top 10 region-to-gene links per gene: 5%| | 26/574 [
Processing: negative r2g, Top 10 region-to-gene links per gene: 5%| | 31/574 [
Processing: negative r2g, Top 10 region-to-gene links per gene: 6%| | 35/574 [
Processing: negative r2g, Top 10 region-to-gene links per gene: 7%| | 39/574 [
Processing: negative r2g, Top 10 region-to-gene links per gene: 7%| | 43/574 [
Processing: negative r2g, Top 10 region-to-gene links per gene: 8%| | 47/574 [
Processing: negative r2g, Top 10 region-to-gene links per gene: 9%| | 51/574 [
Processing: negative r2g, Top 10 region-to-gene links per gene: 10%| | 55/574 [
Processing: negative r2g, Top 10 region-to-gene links per gene: 10%| | 59/574 [
Processing: negative r2g, Top 10 region-to-gene links per gene: 11%| | 63/574 [
Processing: negative r2g, Top 10 region-to-gene links per gene: 12%| | 67/574 [
Processing: negative r2g, Top 10 region-to-gene links per gene: 12%| | 71/574 [
Processing: negative r2g, Top 10 region-to-gene links per gene: 13%|▏| 76/574 [
Processing: negative r2g, Top 10 region-to-gene links per gene: 14%|▏| 80/574 [
Processing: negative r2g, Top 10 region-to-gene links per gene: 15%|▏| 84/574 [
Processing: negative r2g, Top 10 region-to-gene links per gene: 15%|▏| 88/574 [
Processing: negative r2g, Top 10 region-to-gene links per gene: 16%|▏| 91/574 [
Processing: negative r2g, Top 10 region-to-gene links per gene: 16%|▏| 94/574 [
Processing: negative r2g, Top 10 region-to-gene links per gene: 17%|▏| 97/574 [
Processing: negative r2g, Top 10 region-to-gene links per gene: 18%|▏| 101/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 18%|▏| 104/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 19%|▏| 107/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 19%|▏| 111/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 20%|▏| 116/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 21%|▏| 120/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 22%|▏| 124/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 22%|▏| 129/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 23%|▏| 133/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 24%|▏| 137/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 25%|▏| 141/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 25%|▎| 145/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 26%|▎| 149/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 27%|▎| 154/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 28%|▎| 158/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 28%|▎| 162/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 29%|▎| 166/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 29%|▎| 169/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 30%|▎| 172/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 31%|▎| 176/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 31%|▎| 180/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 32%|▎| 185/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 33%|▎| 189/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 34%|▎| 193/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 34%|▎| 197/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 35%|▎| 200/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 36%|▎| 204/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 36%|▎| 208/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 37%|▎| 211/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 37%|▎| 215/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 38%|▍| 218/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 39%|▍| 221/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 39%|▍| 226/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 40%|▍| 230/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 41%|▍| 234/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 41%|▍| 238/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 42%|▍| 242/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 43%|▍| 246/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 44%|▍| 250/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 44%|▍| 254/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 45%|▍| 258/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 46%|▍| 263/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 47%|▍| 267/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 47%|▍| 271/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 48%|▍| 276/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 49%|▍| 281/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 50%|▍| 285/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 50%|▌| 289/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 51%|▌| 293/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 52%|▌| 298/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 53%|▌| 303/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 53%|▌| 307/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 54%|▌| 312/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 55%|▌| 316/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 56%|▌| 320/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 56%|▌| 323/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 57%|▌| 327/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 58%|▌| 331/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 58%|▌| 335/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 59%|▌| 339/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 60%|▌| 344/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 61%|▌| 349/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 61%|▌| 353/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 62%|▌| 357/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 63%|▋| 362/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 64%|▋| 366/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 64%|▋| 370/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 65%|▋| 374/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 66%|▋| 378/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 67%|▋| 382/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 67%|▋| 385/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 68%|▋| 388/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 68%|▋| 393/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 69%|▋| 396/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 70%|▋| 399/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 70%|▋| 404/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 71%|▋| 408/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 72%|▋| 412/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 72%|▋| 416/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 73%|▋| 420/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 74%|▋| 424/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 75%|▋| 428/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 75%|▊| 433/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 76%|▊| 437/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 77%|▊| 441/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 78%|▊| 445/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 78%|▊| 449/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 79%|▊| 453/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 80%|▊| 458/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 80%|▊| 462/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 81%|▊| 466/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 82%|▊| 471/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 83%|▊| 475/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 83%|▊| 479/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 84%|▊| 483/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 85%|▊| 487/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 86%|▊| 491/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 86%|▊| 496/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 87%|▊| 500/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 88%|▉| 504/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 88%|▉| 507/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 89%|▉| 511/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 90%|▉| 515/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 90%|▉| 519/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 91%|▉| 524/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 92%|▉| 529/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 93%|▉| 533/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 94%|▉| 537/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 94%|▉| 542/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 95%|▉| 546/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 96%|▉| 550/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 97%|▉| 554/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 97%|▉| 559/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 98%|▉| 563/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 99%|▉| 567/574
Processing: negative r2g, Top 10 region-to-gene links per gene: 99%|▉| 571/574
36%|███████████████▋ | 5/14 [00:52<01:52, 12.48s/it]
Processing: Top 15 region-to-gene links per gene, negative r2g: 0%| | 0/574 [0
Processing: Top 15 region-to-gene links per gene, negative r2g: 0%| | 2/574 [0
Processing: Top 15 region-to-gene links per gene, negative r2g: 1%| | 4/574 [0
Processing: Top 15 region-to-gene links per gene, negative r2g: 1%| | 7/574 [0
Processing: Top 15 region-to-gene links per gene, negative r2g: 2%| | 10/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 2%| | 13/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 3%| | 16/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 3%| | 19/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 4%| | 22/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 4%| | 25/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 5%| | 28/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 6%| | 32/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 6%| | 35/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 7%| | 38/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 7%| | 41/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 8%| | 44/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 8%| | 47/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 9%| | 50/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 9%| | 53/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 10%| | 56/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 10%| | 59/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 11%| | 62/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 11%| | 65/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 12%| | 68/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 12%| | 71/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 13%|▏| 74/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 13%|▏| 77/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 14%|▏| 80/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 14%|▏| 83/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 15%|▏| 86/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 16%|▏| 89/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 16%|▏| 92/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 17%|▏| 95/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 17%|▏| 98/574 [
Processing: Top 15 region-to-gene links per gene, negative r2g: 18%|▏| 101/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 18%|▏| 104/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 19%|▏| 107/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 19%|▏| 110/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 20%|▏| 113/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 20%|▏| 116/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 21%|▏| 119/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 21%|▏| 122/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 22%|▏| 125/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 22%|▏| 128/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 23%|▏| 132/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 24%|▏| 135/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 24%|▏| 138/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 25%|▏| 141/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 25%|▎| 144/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 26%|▎| 147/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 26%|▎| 150/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 27%|▎| 153/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 27%|▎| 156/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 28%|▎| 159/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 28%|▎| 162/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 29%|▎| 165/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 29%|▎| 168/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 30%|▎| 171/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 30%|▎| 174/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 31%|▎| 177/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 31%|▎| 180/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 32%|▎| 183/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 32%|▎| 186/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 33%|▎| 189/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 33%|▎| 192/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 34%|▎| 195/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 34%|▎| 198/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 35%|▎| 201/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 36%|▎| 204/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 36%|▎| 207/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 37%|▎| 210/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 37%|▎| 213/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 38%|▍| 216/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 38%|▍| 219/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 39%|▍| 222/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 39%|▍| 225/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 40%|▍| 228/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 40%|▍| 231/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 41%|▍| 235/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 41%|▍| 238/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 42%|▍| 241/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 43%|▍| 245/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 43%|▍| 248/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 44%|▍| 251/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 44%|▍| 254/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 45%|▍| 257/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 45%|▍| 260/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 46%|▍| 263/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 46%|▍| 266/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 47%|▍| 269/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 47%|▍| 272/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 48%|▍| 275/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 48%|▍| 278/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 49%|▍| 281/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 49%|▍| 284/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 50%|▌| 287/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 51%|▌| 290/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 51%|▌| 293/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 52%|▌| 296/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 52%|▌| 299/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 53%|▌| 302/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 53%|▌| 305/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 54%|▌| 308/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 54%|▌| 311/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 55%|▌| 314/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 55%|▌| 317/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 56%|▌| 319/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 56%|▌| 322/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 57%|▌| 326/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 57%|▌| 329/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 58%|▌| 332/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 58%|▌| 335/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 59%|▌| 338/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 59%|▌| 341/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 60%|▌| 344/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 60%|▌| 347/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 61%|▌| 350/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 61%|▌| 353/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 62%|▌| 356/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 63%|▋| 359/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 63%|▋| 362/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 64%|▋| 365/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 64%|▋| 368/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 65%|▋| 371/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 65%|▋| 374/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 66%|▋| 377/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 66%|▋| 380/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 67%|▋| 383/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 67%|▋| 386/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 68%|▋| 389/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 68%|▋| 392/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 69%|▋| 395/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 69%|▋| 398/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 70%|▋| 401/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 70%|▋| 404/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 71%|▋| 407/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 71%|▋| 410/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 72%|▋| 413/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 72%|▋| 416/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 73%|▋| 419/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 74%|▋| 422/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 74%|▋| 425/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 75%|▋| 429/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 75%|▊| 432/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 76%|▊| 435/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 76%|▊| 438/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 77%|▊| 441/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 77%|▊| 444/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 78%|▊| 447/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 78%|▊| 450/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 79%|▊| 453/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 79%|▊| 456/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 80%|▊| 459/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 80%|▊| 462/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 81%|▊| 465/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 82%|▊| 469/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 82%|▊| 472/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 83%|▊| 475/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 83%|▊| 478/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 84%|▊| 481/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 84%|▊| 484/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 85%|▊| 487/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 85%|▊| 490/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 86%|▊| 493/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 86%|▊| 496/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 87%|▊| 499/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 87%|▊| 502/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 88%|▉| 505/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 89%|▉| 508/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 89%|▉| 511/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 90%|▉| 514/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 90%|▉| 517/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 91%|▉| 520/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 91%|▉| 523/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 92%|▉| 526/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 92%|▉| 529/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 93%|▉| 532/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 93%|▉| 535/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 94%|▉| 538/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 94%|▉| 542/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 95%|▉| 545/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 95%|▉| 548/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 96%|▉| 551/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 97%|▉| 554/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 97%|▉| 557/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 98%|▉| 560/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 98%|▉| 563/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 99%|▉| 566/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 99%|▉| 569/574
Processing: Top 15 region-to-gene links per gene, negative r2g: 100%|▉| 572/574
43%|██████████████████▊ | 6/14 [01:19<02:19, 17.43s/it]
Processing: negative r2g, BASC binarized: 0%| | 0/574 [00:00<?, ?it/s]
Processing: negative r2g, BASC binarized: 1%| | 6/574 [00:00<00:10, 55.86it/s]
Processing: negative r2g, BASC binarized: 2%| | 14/574 [00:00<00:08, 67.10it/s
Processing: negative r2g, BASC binarized: 4%| | 21/574 [00:00<00:09, 59.87it/s
Processing: negative r2g, BASC binarized: 5%| | 29/574 [00:00<00:08, 65.78it/s
Processing: negative r2g, BASC binarized: 6%| | 36/574 [00:00<00:08, 65.69it/s
Processing: negative r2g, BASC binarized: 7%| | 43/574 [00:00<00:07, 66.88it/s
Processing: negative r2g, BASC binarized: 9%| | 51/574 [00:00<00:07, 69.68it/s
Processing: negative r2g, BASC binarized: 10%| | 59/574 [00:00<00:08, 59.50it/s
Processing: negative r2g, BASC binarized: 11%| | 66/574 [00:01<00:08, 58.25it/s
Processing: negative r2g, BASC binarized: 13%|▏| 74/574 [00:01<00:07, 63.14it/s
Processing: negative r2g, BASC binarized: 14%|▏| 82/574 [00:01<00:07, 65.36it/s
Processing: negative r2g, BASC binarized: 16%|▏| 90/574 [00:01<00:07, 67.73it/s
Processing: negative r2g, BASC binarized: 17%|▏| 98/574 [00:01<00:06, 68.81it/s
Processing: negative r2g, BASC binarized: 18%|▏| 105/574 [00:01<00:07, 63.25it/
Processing: negative r2g, BASC binarized: 20%|▏| 112/574 [00:01<00:07, 59.83it/
Processing: negative r2g, BASC binarized: 21%|▏| 120/574 [00:01<00:07, 63.31it/
Processing: negative r2g, BASC binarized: 22%|▏| 129/574 [00:01<00:06, 68.66it/
Processing: negative r2g, BASC binarized: 24%|▏| 136/574 [00:02<00:07, 59.94it/
Processing: negative r2g, BASC binarized: 25%|▏| 143/574 [00:02<00:06, 61.86it/
Processing: negative r2g, BASC binarized: 26%|▎| 150/574 [00:02<00:07, 58.31it/
Processing: negative r2g, BASC binarized: 28%|▎| 158/574 [00:02<00:06, 62.57it/
Processing: negative r2g, BASC binarized: 29%|▎| 165/574 [00:03<00:27, 14.64it/
Processing: negative r2g, BASC binarized: 30%|▎| 173/574 [00:03<00:20, 19.73it/
Processing: negative r2g, BASC binarized: 31%|▎| 180/574 [00:04<00:15, 24.71it/
Processing: negative r2g, BASC binarized: 32%|▎| 186/574 [00:04<00:13, 29.07it/
Processing: negative r2g, BASC binarized: 33%|▎| 192/574 [00:04<00:12, 31.26it/
Processing: negative r2g, BASC binarized: 34%|▎| 198/574 [00:04<00:10, 36.06it/
Processing: negative r2g, BASC binarized: 36%|▎| 205/574 [00:04<00:09, 40.76it/
Processing: negative r2g, BASC binarized: 37%|▎| 211/574 [00:04<00:08, 43.65it/
Processing: negative r2g, BASC binarized: 38%|▍| 219/574 [00:04<00:07, 50.65it/
Processing: negative r2g, BASC binarized: 39%|▍| 226/574 [00:04<00:06, 54.43it/
Processing: negative r2g, BASC binarized: 41%|▍| 233/574 [00:05<00:06, 52.63it/
Processing: negative r2g, BASC binarized: 42%|▍| 239/574 [00:05<00:06, 54.36it/
Processing: negative r2g, BASC binarized: 43%|▍| 247/574 [00:05<00:05, 58.95it/
Processing: negative r2g, BASC binarized: 44%|▍| 254/574 [00:05<00:05, 56.47it/
Processing: negative r2g, BASC binarized: 45%|▍| 261/574 [00:05<00:05, 59.44it/
Processing: negative r2g, BASC binarized: 47%|▍| 269/574 [00:05<00:04, 63.23it/
Processing: negative r2g, BASC binarized: 48%|▍| 276/574 [00:05<00:05, 58.58it/
Processing: negative r2g, BASC binarized: 49%|▍| 283/574 [00:05<00:04, 58.24it/
Processing: negative r2g, BASC binarized: 51%|▌| 290/574 [00:05<00:04, 59.65it/
Processing: negative r2g, BASC binarized: 52%|▌| 297/574 [00:06<00:04, 60.22it/
Processing: negative r2g, BASC binarized: 53%|▌| 304/574 [00:06<00:04, 59.68it/
Processing: negative r2g, BASC binarized: 54%|▌| 311/574 [00:06<00:05, 52.35it/
Processing: negative r2g, BASC binarized: 55%|▌| 317/574 [00:06<00:05, 45.16it/
Processing: negative r2g, BASC binarized: 56%|▌| 323/574 [00:06<00:05, 47.54it/
Processing: negative r2g, BASC binarized: 57%|▌| 330/574 [00:06<00:04, 52.59it/
Processing: negative r2g, BASC binarized: 59%|▌| 336/574 [00:06<00:04, 54.14it/
Processing: negative r2g, BASC binarized: 60%|▌| 342/574 [00:07<00:04, 50.77it/
Processing: negative r2g, BASC binarized: 61%|▌| 350/574 [00:07<00:03, 57.99it/
Processing: negative r2g, BASC binarized: 62%|▌| 357/574 [00:07<00:03, 60.64it/
Processing: negative r2g, BASC binarized: 63%|▋| 364/574 [00:07<00:03, 56.14it/
Processing: negative r2g, BASC binarized: 64%|▋| 370/574 [00:07<00:03, 54.49it/
Processing: negative r2g, BASC binarized: 66%|▋| 376/574 [00:07<00:04, 48.88it/
Processing: negative r2g, BASC binarized: 67%|▋| 382/574 [00:07<00:03, 48.43it/
Processing: negative r2g, BASC binarized: 67%|▋| 387/574 [00:07<00:03, 48.45it/
Processing: negative r2g, BASC binarized: 68%|▋| 393/574 [00:08<00:03, 48.94it/
Processing: negative r2g, BASC binarized: 69%|▋| 398/574 [00:08<00:03, 47.91it/
Processing: negative r2g, BASC binarized: 71%|▋| 406/574 [00:08<00:03, 55.73it/
Processing: negative r2g, BASC binarized: 72%|▋| 413/574 [00:08<00:02, 58.19it/
Processing: negative r2g, BASC binarized: 73%|▋| 419/574 [00:08<00:02, 53.78it/
Processing: negative r2g, BASC binarized: 74%|▋| 427/574 [00:08<00:02, 59.37it/
Processing: negative r2g, BASC binarized: 76%|▊| 434/574 [00:08<00:02, 59.54it/
Processing: negative r2g, BASC binarized: 77%|▊| 441/574 [00:08<00:02, 58.40it/
Processing: negative r2g, BASC binarized: 78%|▊| 447/574 [00:08<00:02, 53.38it/
Processing: negative r2g, BASC binarized: 79%|▊| 453/574 [00:09<00:02, 53.33it/
Processing: negative r2g, BASC binarized: 80%|▊| 459/574 [00:09<00:02, 54.98it/
Processing: negative r2g, BASC binarized: 81%|▊| 466/574 [00:09<00:01, 57.78it/
Processing: negative r2g, BASC binarized: 83%|▊| 475/574 [00:09<00:01, 63.75it/
Processing: negative r2g, BASC binarized: 84%|▊| 482/574 [00:09<00:01, 64.19it/
Processing: negative r2g, BASC binarized: 85%|▊| 489/574 [00:09<00:01, 62.70it/
Processing: negative r2g, BASC binarized: 86%|▊| 496/574 [00:09<00:01, 60.04it/
Processing: negative r2g, BASC binarized: 88%|▉| 503/574 [00:09<00:01, 57.84it/
Processing: negative r2g, BASC binarized: 89%|▉| 509/574 [00:09<00:01, 58.03it/
Processing: negative r2g, BASC binarized: 90%|▉| 517/574 [00:10<00:00, 61.73it/
Processing: negative r2g, BASC binarized: 91%|▉| 524/574 [00:10<00:00, 61.45it/
Processing: negative r2g, BASC binarized: 93%|▉| 531/574 [00:10<00:00, 63.49it/
Processing: negative r2g, BASC binarized: 94%|▉| 538/574 [00:10<00:00, 61.81it/
Processing: negative r2g, BASC binarized: 95%|▉| 545/574 [00:10<00:00, 63.04it/
Processing: negative r2g, BASC binarized: 96%|▉| 552/574 [00:10<00:00, 64.44it/
Processing: negative r2g, BASC binarized: 97%|▉| 559/574 [00:10<00:00, 64.24it/
Processing: negative r2g, BASC binarized: 99%|▉| 566/574 [00:10<00:00, 61.10it/
Processing: negative r2g, BASC binarized: 100%|▉| 573/574 [00:11<00:00, 57.92it/
50%|██████████████████████ | 7/14 [01:41<02:13, 19.00s/it]
Processing: 0.85 quantile, positive r2g: 0%| | 0/574 [00:00<?, ?it/s]
Processing: 0.85 quantile, positive r2g: 1%| | 4/574 [00:00<00:15, 37.94it/s]
Processing: 0.85 quantile, positive r2g: 2%| | 9/574 [00:00<00:13, 43.11it/s]
Processing: 0.85 quantile, positive r2g: 2%| | 14/574 [00:00<00:13, 41.86it/s]
Processing: 0.85 quantile, positive r2g: 3%| | 19/574 [00:00<00:14, 37.22it/s]
Processing: 0.85 quantile, positive r2g: 4%| | 24/574 [00:00<00:13, 40.29it/s]
Processing: 0.85 quantile, positive r2g: 5%| | 30/574 [00:00<00:12, 42.53it/s]
Processing: 0.85 quantile, positive r2g: 6%| | 35/574 [00:00<00:13, 40.71it/s]
Processing: 0.85 quantile, positive r2g: 7%| | 41/574 [00:00<00:12, 43.92it/s]
Processing: 0.85 quantile, positive r2g: 8%| | 46/574 [00:01<00:12, 41.16it/s]
Processing: 0.85 quantile, positive r2g: 9%| | 51/574 [00:01<00:13, 39.76it/s]
Processing: 0.85 quantile, positive r2g: 10%| | 56/574 [00:01<00:14, 36.76it/s]
Processing: 0.85 quantile, positive r2g: 11%| | 61/574 [00:01<00:12, 39.76it/s]
Processing: 0.85 quantile, positive r2g: 12%| | 67/574 [00:01<00:12, 42.25it/s]
Processing: 0.85 quantile, positive r2g: 13%|▏| 72/574 [00:01<00:13, 38.34it/s]
Processing: 0.85 quantile, positive r2g: 14%|▏| 78/574 [00:01<00:11, 42.27it/s]
Processing: 0.85 quantile, positive r2g: 15%|▏| 84/574 [00:02<00:10, 44.78it/s]
Processing: 0.85 quantile, positive r2g: 16%|▏| 89/574 [00:02<00:11, 42.35it/s]
Processing: 0.85 quantile, positive r2g: 16%|▏| 94/574 [00:02<00:10, 43.86it/s]
Processing: 0.85 quantile, positive r2g: 17%|▏| 99/574 [00:02<00:12, 37.65it/s]
Processing: 0.85 quantile, positive r2g: 18%|▏| 104/574 [00:02<00:11, 39.40it/s
Processing: 0.85 quantile, positive r2g: 19%|▏| 110/574 [00:02<00:10, 42.51it/s
Processing: 0.85 quantile, positive r2g: 20%|▏| 115/574 [00:02<00:10, 42.20it/s
Processing: 0.85 quantile, positive r2g: 21%|▏| 120/574 [00:02<00:11, 39.23it/s
Processing: 0.85 quantile, positive r2g: 22%|▏| 126/574 [00:03<00:10, 42.96it/s
Processing: 0.85 quantile, positive r2g: 23%|▏| 131/574 [00:03<00:10, 41.89it/s
Processing: 0.85 quantile, positive r2g: 24%|▏| 136/574 [00:03<00:10, 42.24it/s
Processing: 0.85 quantile, positive r2g: 25%|▏| 141/574 [00:03<00:10, 40.20it/s
Processing: 0.85 quantile, positive r2g: 25%|▎| 146/574 [00:03<00:11, 38.25it/s
Processing: 0.85 quantile, positive r2g: 26%|▎| 151/574 [00:03<00:10, 38.51it/s
Processing: 0.85 quantile, positive r2g: 27%|▎| 156/574 [00:03<00:10, 41.02it/s
Processing: 0.85 quantile, positive r2g: 28%|▎| 161/574 [00:03<00:09, 41.73it/s
Processing: 0.85 quantile, positive r2g: 29%|▎| 166/574 [00:04<00:10, 37.16it/s
Processing: 0.85 quantile, positive r2g: 30%|▎| 171/574 [00:04<00:10, 39.52it/s
Processing: 0.85 quantile, positive r2g: 31%|▎| 176/574 [00:04<00:09, 41.70it/s
Processing: 0.85 quantile, positive r2g: 32%|▎| 181/574 [00:04<00:09, 40.30it/s
Processing: 0.85 quantile, positive r2g: 32%|▎| 186/574 [00:04<00:10, 38.38it/s
Processing: 0.85 quantile, positive r2g: 33%|▎| 190/574 [00:04<00:10, 35.54it/s
Processing: 0.85 quantile, positive r2g: 34%|▎| 194/574 [00:04<00:10, 36.23it/s
Processing: 0.85 quantile, positive r2g: 35%|▎| 199/574 [00:04<00:09, 39.21it/s
Processing: 0.85 quantile, positive r2g: 36%|▎| 204/574 [00:05<00:09, 38.58it/s
Processing: 0.85 quantile, positive r2g: 36%|▎| 208/574 [00:06<00:40, 9.08it/s
Processing: 0.85 quantile, positive r2g: 37%|▎| 212/574 [00:06<00:31, 11.52it/s
Processing: 0.85 quantile, positive r2g: 38%|▍| 217/574 [00:06<00:23, 15.22it/s
Processing: 0.85 quantile, positive r2g: 39%|▍| 222/574 [00:06<00:18, 19.06it/s
Processing: 0.85 quantile, positive r2g: 40%|▍| 227/574 [00:06<00:14, 23.58it/s
Processing: 0.85 quantile, positive r2g: 40%|▍| 232/574 [00:07<00:12, 27.54it/s
Processing: 0.85 quantile, positive r2g: 41%|▍| 237/574 [00:07<00:11, 28.78it/s
Processing: 0.85 quantile, positive r2g: 42%|▍| 242/574 [00:07<00:10, 32.33it/s
Processing: 0.85 quantile, positive r2g: 43%|▍| 248/574 [00:07<00:08, 36.58it/s
Processing: 0.85 quantile, positive r2g: 44%|▍| 253/574 [00:07<00:09, 34.73it/s
Processing: 0.85 quantile, positive r2g: 45%|▍| 257/574 [00:07<00:09, 35.04it/s
Processing: 0.85 quantile, positive r2g: 46%|▍| 263/574 [00:07<00:07, 39.01it/s
Processing: 0.85 quantile, positive r2g: 47%|▍| 268/574 [00:07<00:08, 36.79it/s
Processing: 0.85 quantile, positive r2g: 48%|▍| 273/574 [00:08<00:07, 38.43it/s
Processing: 0.85 quantile, positive r2g: 48%|▍| 278/574 [00:08<00:08, 36.84it/s
Processing: 0.85 quantile, positive r2g: 49%|▍| 282/574 [00:08<00:07, 36.58it/s
Processing: 0.85 quantile, positive r2g: 50%|▌| 287/574 [00:08<00:07, 39.44it/s
Processing: 0.85 quantile, positive r2g: 51%|▌| 292/574 [00:08<00:07, 38.06it/s
Processing: 0.85 quantile, positive r2g: 52%|▌| 296/574 [00:08<00:07, 35.42it/s
Processing: 0.85 quantile, positive r2g: 52%|▌| 301/574 [00:08<00:07, 37.55it/s
Processing: 0.85 quantile, positive r2g: 53%|▌| 305/574 [00:08<00:07, 36.52it/s
Processing: 0.85 quantile, positive r2g: 54%|▌| 309/574 [00:09<00:08, 33.03it/s
Processing: 0.85 quantile, positive r2g: 55%|▌| 313/574 [00:09<00:07, 32.75it/s
Processing: 0.85 quantile, positive r2g: 55%|▌| 317/574 [00:09<00:07, 33.15it/s
Processing: 0.85 quantile, positive r2g: 56%|▌| 321/574 [00:09<00:07, 32.09it/s
Processing: 0.85 quantile, positive r2g: 57%|▌| 325/574 [00:09<00:07, 33.59it/s
Processing: 0.85 quantile, positive r2g: 57%|▌| 329/574 [00:09<00:07, 31.78it/s
Processing: 0.85 quantile, positive r2g: 58%|▌| 334/574 [00:09<00:06, 35.47it/s
Processing: 0.85 quantile, positive r2g: 59%|▌| 338/574 [00:09<00:06, 36.25it/s
Processing: 0.85 quantile, positive r2g: 60%|▌| 342/574 [00:10<00:06, 37.09it/s
Processing: 0.85 quantile, positive r2g: 60%|▌| 346/574 [00:10<00:06, 36.58it/s
Processing: 0.85 quantile, positive r2g: 61%|▌| 350/574 [00:10<00:06, 34.00it/s
Processing: 0.85 quantile, positive r2g: 62%|▌| 354/574 [00:10<00:06, 34.88it/s
Processing: 0.85 quantile, positive r2g: 63%|▋| 359/574 [00:10<00:05, 38.82it/s
Processing: 0.85 quantile, positive r2g: 63%|▋| 364/574 [00:10<00:05, 40.36it/s
Processing: 0.85 quantile, positive r2g: 64%|▋| 369/574 [00:10<00:06, 34.09it/s
Processing: 0.85 quantile, positive r2g: 65%|▋| 374/574 [00:10<00:05, 36.48it/s
Processing: 0.85 quantile, positive r2g: 66%|▋| 378/574 [00:11<00:05, 34.52it/s
Processing: 0.85 quantile, positive r2g: 67%|▋| 382/574 [00:11<00:05, 35.84it/s
Processing: 0.85 quantile, positive r2g: 67%|▋| 386/574 [00:11<00:05, 35.39it/s
Processing: 0.85 quantile, positive r2g: 68%|▋| 390/574 [00:11<00:05, 33.75it/s
Processing: 0.85 quantile, positive r2g: 69%|▋| 394/574 [00:11<00:05, 35.21it/s
Processing: 0.85 quantile, positive r2g: 69%|▋| 398/574 [00:11<00:04, 35.87it/s
Processing: 0.85 quantile, positive r2g: 70%|▋| 402/574 [00:11<00:04, 36.11it/s
Processing: 0.85 quantile, positive r2g: 71%|▋| 407/574 [00:11<00:04, 39.28it/s
Processing: 0.85 quantile, positive r2g: 72%|▋| 412/574 [00:11<00:03, 40.89it/s
Processing: 0.85 quantile, positive r2g: 73%|▋| 417/574 [00:12<00:04, 37.03it/s
Processing: 0.85 quantile, positive r2g: 74%|▋| 422/574 [00:12<00:03, 38.69it/s
Processing: 0.85 quantile, positive r2g: 74%|▋| 426/574 [00:12<00:03, 37.56it/s
Processing: 0.85 quantile, positive r2g: 75%|▋| 430/574 [00:12<00:03, 38.00it/s
Processing: 0.85 quantile, positive r2g: 76%|▊| 435/574 [00:12<00:03, 38.69it/s
Processing: 0.85 quantile, positive r2g: 76%|▊| 439/574 [00:12<00:03, 37.56it/s
Processing: 0.85 quantile, positive r2g: 77%|▊| 443/574 [00:12<00:03, 37.98it/s
Processing: 0.85 quantile, positive r2g: 78%|▊| 447/574 [00:12<00:03, 34.97it/s
Processing: 0.85 quantile, positive r2g: 79%|▊| 451/574 [00:13<00:03, 36.15it/s
Processing: 0.85 quantile, positive r2g: 79%|▊| 456/574 [00:13<00:03, 38.84it/s
Processing: 0.85 quantile, positive r2g: 80%|▊| 461/574 [00:13<00:02, 41.21it/s
Processing: 0.85 quantile, positive r2g: 81%|▊| 467/574 [00:13<00:02, 43.24it/s
Processing: 0.85 quantile, positive r2g: 82%|▊| 472/574 [00:13<00:02, 37.89it/s
Processing: 0.85 quantile, positive r2g: 83%|▊| 476/574 [00:13<00:02, 37.75it/s
Processing: 0.85 quantile, positive r2g: 84%|▊| 481/574 [00:13<00:02, 40.49it/s
Processing: 0.85 quantile, positive r2g: 85%|▊| 486/574 [00:13<00:02, 41.72it/s
Processing: 0.85 quantile, positive r2g: 86%|▊| 491/574 [00:13<00:01, 42.13it/s
Processing: 0.85 quantile, positive r2g: 86%|▊| 496/574 [00:14<00:02, 37.40it/s
Processing: 0.85 quantile, positive r2g: 87%|▊| 501/574 [00:14<00:01, 40.44it/s
Processing: 0.85 quantile, positive r2g: 88%|▉| 506/574 [00:14<00:01, 40.25it/s
Processing: 0.85 quantile, positive r2g: 89%|▉| 511/574 [00:14<00:01, 41.61it/s
Processing: 0.85 quantile, positive r2g: 90%|▉| 516/574 [00:14<00:01, 42.98it/s
Processing: 0.85 quantile, positive r2g: 91%|▉| 521/574 [00:14<00:01, 38.88it/s
Processing: 0.85 quantile, positive r2g: 92%|▉| 527/574 [00:14<00:01, 42.63it/s
Processing: 0.85 quantile, positive r2g: 93%|▉| 532/574 [00:15<00:01, 37.70it/s
Processing: 0.85 quantile, positive r2g: 94%|▉| 537/574 [00:15<00:00, 39.96it/s
Processing: 0.85 quantile, positive r2g: 94%|▉| 542/574 [00:15<00:00, 42.28it/s
Processing: 0.85 quantile, positive r2g: 95%|▉| 547/574 [00:15<00:00, 42.70it/s
Processing: 0.85 quantile, positive r2g: 96%|▉| 552/574 [00:15<00:00, 40.16it/s
Processing: 0.85 quantile, positive r2g: 97%|▉| 557/574 [00:15<00:00, 39.75it/s
Processing: 0.85 quantile, positive r2g: 98%|▉| 562/574 [00:15<00:00, 41.15it/s
Processing: 0.85 quantile, positive r2g: 99%|▉| 567/574 [00:15<00:00, 43.23it/s
Processing: 0.85 quantile, positive r2g: 100%|▉| 572/574 [00:15<00:00, 44.28it/s
57%|█████████████████████████▏ | 8/14 [01:59<01:52, 18.73s/it]
Processing: 0.9 quantile, positive r2g: 0%| | 0/574 [00:00<?, ?it/s]
Processing: 0.9 quantile, positive r2g: 1%| | 7/574 [00:00<00:08, 63.12it/s]
Processing: 0.9 quantile, positive r2g: 2%| | 14/574 [00:00<00:09, 59.13it/s]
Processing: 0.9 quantile, positive r2g: 4%| | 21/574 [00:00<00:09, 61.02it/s]
Processing: 0.9 quantile, positive r2g: 5%| | 28/574 [00:00<00:09, 56.00it/s]
Processing: 0.9 quantile, positive r2g: 6%| | 34/574 [00:00<00:09, 54.61it/s]
Processing: 0.9 quantile, positive r2g: 7%|▏ | 41/574 [00:00<00:09, 58.79it/s]
Processing: 0.9 quantile, positive r2g: 8%|▏ | 48/574 [00:00<00:08, 60.40it/s]
Processing: 0.9 quantile, positive r2g: 10%|▏ | 55/574 [00:00<00:10, 51.13it/s]
Processing: 0.9 quantile, positive r2g: 11%|▏ | 62/574 [00:01<00:09, 54.33it/s]
Processing: 0.9 quantile, positive r2g: 12%|▏ | 70/574 [00:01<00:08, 59.54it/s]
Processing: 0.9 quantile, positive r2g: 14%|▎ | 78/574 [00:01<00:07, 64.25it/s]
Processing: 0.9 quantile, positive r2g: 15%|▎ | 85/574 [00:01<00:08, 58.99it/s]
Processing: 0.9 quantile, positive r2g: 16%|▎ | 93/574 [00:01<00:07, 62.85it/s]
Processing: 0.9 quantile, positive r2g: 17%|▏| 100/574 [00:01<00:07, 62.55it/s]
Processing: 0.9 quantile, positive r2g: 19%|▏| 107/574 [00:01<00:07, 60.84it/s]
Processing: 0.9 quantile, positive r2g: 20%|▏| 115/574 [00:01<00:07, 64.82it/s]
Processing: 0.9 quantile, positive r2g: 21%|▏| 122/574 [00:02<00:06, 65.51it/s]
Processing: 0.9 quantile, positive r2g: 22%|▏| 129/574 [00:02<00:06, 65.38it/s]
Processing: 0.9 quantile, positive r2g: 24%|▏| 136/574 [00:02<00:06, 64.37it/s]
Processing: 0.9 quantile, positive r2g: 25%|▎| 144/574 [00:02<00:06, 66.55it/s]
Processing: 0.9 quantile, positive r2g: 26%|▎| 152/574 [00:02<00:06, 68.25it/s]
Processing: 0.9 quantile, positive r2g: 28%|▎| 159/574 [00:02<00:06, 65.25it/s]
Processing: 0.9 quantile, positive r2g: 29%|▎| 166/574 [00:02<00:06, 58.45it/s]
Processing: 0.9 quantile, positive r2g: 30%|▎| 173/574 [00:02<00:06, 59.39it/s]
Processing: 0.9 quantile, positive r2g: 32%|▎| 181/574 [00:02<00:06, 63.05it/s]
Processing: 0.9 quantile, positive r2g: 33%|▎| 188/574 [00:03<00:06, 63.21it/s]
Processing: 0.9 quantile, positive r2g: 34%|▎| 195/574 [00:03<00:06, 54.73it/s]
Processing: 0.9 quantile, positive r2g: 35%|▎| 201/574 [00:03<00:07, 52.77it/s]
Processing: 0.9 quantile, positive r2g: 36%|▎| 209/574 [00:03<00:06, 57.46it/s]
Processing: 0.9 quantile, positive r2g: 38%|▍| 216/574 [00:03<00:05, 60.64it/s]
Processing: 0.9 quantile, positive r2g: 39%|▍| 223/574 [00:03<00:05, 61.47it/s]
Processing: 0.9 quantile, positive r2g: 40%|▍| 230/574 [00:03<00:05, 59.04it/s]
Processing: 0.9 quantile, positive r2g: 41%|▍| 237/574 [00:03<00:05, 60.54it/s]
Processing: 0.9 quantile, positive r2g: 43%|▍| 244/574 [00:04<00:05, 62.84it/s]
Processing: 0.9 quantile, positive r2g: 44%|▍| 251/574 [00:04<00:05, 59.87it/s]
Processing: 0.9 quantile, positive r2g: 45%|▍| 258/574 [00:04<00:05, 59.42it/s]
Processing: 0.9 quantile, positive r2g: 46%|▍| 265/574 [00:04<00:05, 59.76it/s]
Processing: 0.9 quantile, positive r2g: 47%|▍| 272/574 [00:04<00:04, 60.96it/s]
Processing: 0.9 quantile, positive r2g: 49%|▍| 280/574 [00:04<00:04, 64.69it/s]
Processing: 0.9 quantile, positive r2g: 50%|▌| 287/574 [00:04<00:04, 61.56it/s]
Processing: 0.9 quantile, positive r2g: 51%|▌| 295/574 [00:04<00:04, 64.84it/s]
Processing: 0.9 quantile, positive r2g: 53%|▌| 303/574 [00:04<00:04, 67.73it/s]
Processing: 0.9 quantile, positive r2g: 54%|▌| 310/574 [00:05<00:04, 64.73it/s]
Processing: 0.9 quantile, positive r2g: 55%|▌| 317/574 [00:05<00:04, 53.42it/s]
Processing: 0.9 quantile, positive r2g: 56%|▌| 324/574 [00:05<00:04, 57.20it/s]
Processing: 0.9 quantile, positive r2g: 58%|▌| 331/574 [00:05<00:04, 60.23it/s]
Processing: 0.9 quantile, positive r2g: 59%|▌| 338/574 [00:05<00:03, 59.75it/s]
Processing: 0.9 quantile, positive r2g: 60%|▌| 345/574 [00:05<00:04, 55.14it/s]
Processing: 0.9 quantile, positive r2g: 61%|▌| 353/574 [00:05<00:03, 59.59it/s]
Processing: 0.9 quantile, positive r2g: 63%|▋| 360/574 [00:05<00:03, 59.90it/s]
Processing: 0.9 quantile, positive r2g: 64%|▋| 367/574 [00:06<00:03, 53.10it/s]
Processing: 0.9 quantile, positive r2g: 65%|▋| 373/574 [00:06<00:03, 53.04it/s]
Processing: 0.9 quantile, positive r2g: 66%|▋| 380/574 [00:06<00:03, 54.01it/s]
Processing: 0.9 quantile, positive r2g: 67%|▋| 386/574 [00:06<00:03, 52.57it/s]
Processing: 0.9 quantile, positive r2g: 68%|▋| 392/574 [00:06<00:03, 53.22it/s]
Processing: 0.9 quantile, positive r2g: 69%|▋| 398/574 [00:06<00:03, 48.88it/s]
Processing: 0.9 quantile, positive r2g: 71%|▋| 405/574 [00:06<00:03, 53.57it/s]
Processing: 0.9 quantile, positive r2g: 72%|▋| 413/574 [00:06<00:02, 58.98it/s]
Processing: 0.9 quantile, positive r2g: 73%|▋| 420/574 [00:07<00:02, 55.07it/s]
Processing: 0.9 quantile, positive r2g: 74%|▋| 426/574 [00:07<00:02, 53.94it/s]
Processing: 0.9 quantile, positive r2g: 76%|▊| 434/574 [00:07<00:02, 59.05it/s]
Processing: 0.9 quantile, positive r2g: 77%|▊| 441/574 [00:07<00:02, 61.76it/s]
Processing: 0.9 quantile, positive r2g: 78%|▊| 448/574 [00:07<00:02, 61.26it/s]
Processing: 0.9 quantile, positive r2g: 79%|▊| 455/574 [00:07<00:02, 53.31it/s]
Processing: 0.9 quantile, positive r2g: 80%|▊| 461/574 [00:07<00:02, 54.49it/s]
Processing: 0.9 quantile, positive r2g: 82%|▊| 469/574 [00:07<00:01, 61.04it/s]
Processing: 0.9 quantile, positive r2g: 83%|▊| 476/574 [00:08<00:01, 60.20it/s]
Processing: 0.9 quantile, positive r2g: 84%|▊| 483/574 [00:08<00:01, 56.57it/s]
Processing: 0.9 quantile, positive r2g: 86%|▊| 491/574 [00:08<00:01, 61.64it/s]
Processing: 0.9 quantile, positive r2g: 87%|▊| 498/574 [00:08<00:01, 60.75it/s]
Processing: 0.9 quantile, positive r2g: 88%|▉| 505/574 [00:08<00:01, 57.38it/s]
Processing: 0.9 quantile, positive r2g: 89%|▉| 511/574 [00:08<00:01, 57.47it/s]
Processing: 0.9 quantile, positive r2g: 90%|▉| 519/574 [00:08<00:00, 61.30it/s]
Processing: 0.9 quantile, positive r2g: 92%|▉| 526/574 [00:08<00:00, 59.34it/s]
Processing: 0.9 quantile, positive r2g: 93%|▉| 532/574 [00:09<00:00, 55.86it/s]
Processing: 0.9 quantile, positive r2g: 94%|▉| 540/574 [00:09<00:00, 60.66it/s]
Processing: 0.9 quantile, positive r2g: 95%|▉| 548/574 [00:09<00:00, 64.97it/s]
Processing: 0.9 quantile, positive r2g: 97%|▉| 555/574 [00:09<00:00, 58.57it/s]
Processing: 0.9 quantile, positive r2g: 98%|▉| 562/574 [00:09<00:00, 58.33it/s]
Processing: 0.9 quantile, positive r2g: 99%|▉| 569/574 [00:09<00:00, 61.18it/s]
64%|████████████████████████████▎ | 9/14 [02:10<01:20, 16.19s/it]
Processing: 0.95 quantile, positive r2g: 0%| | 0/574 [00:00<?, ?it/s]
Processing: 0.95 quantile, positive r2g: 2%| | 12/574 [00:00<00:04, 113.35it/s
Processing: 0.95 quantile, positive r2g: 4%| | 24/574 [00:00<00:04, 111.93it/s
Processing: 0.95 quantile, positive r2g: 6%| | 36/574 [00:00<00:04, 108.81it/s
Processing: 0.95 quantile, positive r2g: 8%| | 48/574 [00:00<00:04, 109.96it/s
Processing: 0.95 quantile, positive r2g: 10%| | 60/574 [00:00<00:04, 111.59it/s
Processing: 0.95 quantile, positive r2g: 13%|▏| 72/574 [00:00<00:04, 113.87it/s
Processing: 0.95 quantile, positive r2g: 15%|▏| 84/574 [00:00<00:04, 111.34it/s
Processing: 0.95 quantile, positive r2g: 17%|▏| 97/574 [00:00<00:04, 115.02it/s
Processing: 0.95 quantile, positive r2g: 19%|▏| 109/574 [00:00<00:04, 110.84it/
Processing: 0.95 quantile, positive r2g: 21%|▏| 121/574 [00:01<00:04, 112.30it/
Processing: 0.95 quantile, positive r2g: 23%|▏| 133/574 [00:01<00:03, 113.36it/
Processing: 0.95 quantile, positive r2g: 25%|▎| 145/574 [00:01<00:03, 113.29it/
Processing: 0.95 quantile, positive r2g: 28%|▎| 158/574 [00:01<00:03, 116.82it/
Processing: 0.95 quantile, positive r2g: 30%|▎| 170/574 [00:01<00:03, 106.73it/
Processing: 0.95 quantile, positive r2g: 32%|▎| 182/574 [00:01<00:03, 108.52it/
Processing: 0.95 quantile, positive r2g: 34%|▎| 193/574 [00:01<00:03, 106.79it/
Processing: 0.95 quantile, positive r2g: 36%|▎| 205/574 [00:01<00:03, 107.49it/
Processing: 0.95 quantile, positive r2g: 38%|▍| 216/574 [00:01<00:03, 106.85it/
Processing: 0.95 quantile, positive r2g: 40%|▍| 228/574 [00:02<00:03, 108.09it/
Processing: 0.95 quantile, positive r2g: 42%|▍| 239/574 [00:02<00:03, 106.20it/
Processing: 0.95 quantile, positive r2g: 44%|▍| 250/574 [00:02<00:03, 106.65it/
Processing: 0.95 quantile, positive r2g: 46%|▍| 262/574 [00:02<00:02, 109.22it/
Processing: 0.95 quantile, positive r2g: 48%|▍| 274/574 [00:02<00:02, 110.68it/
Processing: 0.95 quantile, positive r2g: 50%|▍| 286/574 [00:02<00:02, 112.52it/
Processing: 0.95 quantile, positive r2g: 52%|▌| 298/574 [00:02<00:02, 113.53it/
Processing: 0.95 quantile, positive r2g: 54%|▌| 310/574 [00:02<00:02, 110.35it/
Processing: 0.95 quantile, positive r2g: 56%|▌| 322/574 [00:02<00:02, 102.53it/
Processing: 0.95 quantile, positive r2g: 58%|▌| 334/574 [00:03<00:02, 105.64it/
Processing: 0.95 quantile, positive r2g: 60%|▌| 345/574 [00:03<00:02, 105.33it/
Processing: 0.95 quantile, positive r2g: 62%|▌| 357/574 [00:03<00:01, 109.25it/
Processing: 0.95 quantile, positive r2g: 64%|▋| 369/574 [00:03<00:02, 99.64it/s
Processing: 0.95 quantile, positive r2g: 66%|▋| 380/574 [00:03<00:01, 98.56it/s
Processing: 0.95 quantile, positive r2g: 68%|▋| 392/574 [00:03<00:01, 102.32it/
Processing: 0.95 quantile, positive r2g: 70%|▋| 403/574 [00:03<00:01, 102.88it/
Processing: 0.95 quantile, positive r2g: 72%|▋| 415/574 [00:03<00:01, 107.12it/
Processing: 0.95 quantile, positive r2g: 74%|▋| 427/574 [00:03<00:01, 110.45it/
Processing: 0.95 quantile, positive r2g: 76%|▊| 439/574 [00:04<00:01, 107.35it/
Processing: 0.95 quantile, positive r2g: 78%|▊| 450/574 [00:04<00:01, 104.09it/
Processing: 0.95 quantile, positive r2g: 80%|▊| 461/574 [00:04<00:01, 104.28it/
Processing: 0.95 quantile, positive r2g: 83%|▊| 475/574 [00:04<00:00, 112.03it/
Processing: 0.95 quantile, positive r2g: 85%|▊| 487/574 [00:04<00:00, 110.72it/
Processing: 0.95 quantile, positive r2g: 87%|▊| 499/574 [00:04<00:00, 112.28it/
Processing: 0.95 quantile, positive r2g: 89%|▉| 511/574 [00:04<00:00, 113.71it/
Processing: 0.95 quantile, positive r2g: 91%|▉| 524/574 [00:04<00:00, 116.33it/
Processing: 0.95 quantile, positive r2g: 93%|▉| 536/574 [00:04<00:00, 111.43it/
Processing: 0.95 quantile, positive r2g: 96%|▉| 549/574 [00:05<00:00, 114.83it/
Processing: 0.95 quantile, positive r2g: 98%|▉| 561/574 [00:05<00:00, 114.90it/
Processing: 0.95 quantile, positive r2g: 100%|▉| 573/574 [00:05<00:00, 114.86it/
71%|██████████████████████████████▋ | 10/14 [02:16<00:52, 13.10s/it]
Processing: Top 5 region-to-gene links per gene, positive r2g: 0%| | 0/574 [00
Processing: Top 5 region-to-gene links per gene, positive r2g: 1%| | 4/574 [00
Processing: Top 5 region-to-gene links per gene, positive r2g: 2%| | 10/574 [0
Processing: Top 5 region-to-gene links per gene, positive r2g: 3%| | 17/574 [0
Processing: Top 5 region-to-gene links per gene, positive r2g: 4%| | 24/574 [0
Processing: Top 5 region-to-gene links per gene, positive r2g: 5%| | 30/574 [0
Processing: Top 5 region-to-gene links per gene, positive r2g: 6%| | 36/574 [0
Processing: Top 5 region-to-gene links per gene, positive r2g: 7%| | 42/574 [0
Processing: Top 5 region-to-gene links per gene, positive r2g: 9%| | 49/574 [0
Processing: Top 5 region-to-gene links per gene, positive r2g: 10%| | 56/574 [0
Processing: Top 5 region-to-gene links per gene, positive r2g: 11%| | 63/574 [0
Processing: Top 5 region-to-gene links per gene, positive r2g: 12%| | 71/574 [0
Processing: Top 5 region-to-gene links per gene, positive r2g: 14%|▏| 79/574 [0
Processing: Top 5 region-to-gene links per gene, positive r2g: 15%|▏| 86/574 [0
Processing: Top 5 region-to-gene links per gene, positive r2g: 16%|▏| 92/574 [0
Processing: Top 5 region-to-gene links per gene, positive r2g: 17%|▏| 99/574 [0
Processing: Top 5 region-to-gene links per gene, positive r2g: 18%|▏| 106/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 20%|▏| 113/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 21%|▏| 120/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 22%|▏| 128/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 24%|▏| 135/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 25%|▏| 142/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 26%|▎| 149/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 27%|▎| 157/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 29%|▎| 164/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 30%|▎| 171/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 31%|▎| 178/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 32%|▎| 185/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 33%|▎| 191/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 34%|▎| 198/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 36%|▎| 205/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 37%|▎| 212/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 38%|▍| 218/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 39%|▍| 225/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 40%|▍| 232/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 42%|▍| 239/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 43%|▍| 246/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 44%|▍| 253/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 45%|▍| 260/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 47%|▍| 267/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 48%|▍| 273/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 49%|▍| 280/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 50%|▌| 287/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 51%|▌| 295/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 53%|▌| 302/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 54%|▌| 308/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 55%|▌| 315/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 56%|▌| 321/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 57%|▌| 327/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 58%|▌| 334/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 59%|▌| 340/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 60%|▌| 346/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 61%|▌| 352/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 62%|▌| 358/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 64%|▋| 365/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 65%|▋| 371/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 66%|▋| 377/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 67%|▋| 383/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 68%|▋| 390/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 69%|▋| 396/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 70%|▋| 402/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 71%|▋| 409/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 72%|▋| 415/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 73%|▋| 421/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 74%|▋| 427/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 76%|▊| 434/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 77%|▊| 441/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 78%|▊| 448/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 79%|▊| 455/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 80%|▊| 460/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 81%|▊| 466/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 82%|▊| 473/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 83%|▊| 479/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 85%|▊| 486/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 86%|▊| 492/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 87%|▊| 498/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 88%|▉| 505/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 89%|▉| 512/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 90%|▉| 518/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 91%|▉| 525/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 93%|▉| 532/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 94%|▉| 538/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 95%|▉| 546/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 97%|▉| 554/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 98%|▉| 561/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 99%|▉| 568/574 [
Processing: Top 5 region-to-gene links per gene, positive r2g: 100%|█| 574/574 [
79%|█████████████████████████████████▊ | 11/14 [02:28<00:38, 12.91s/it]
Processing: positive r2g, Top 10 region-to-gene links per gene: 0%| | 0/574 [0
Processing: positive r2g, Top 10 region-to-gene links per gene: 1%| | 3/574 [0
Processing: positive r2g, Top 10 region-to-gene links per gene: 1%| | 7/574 [0
Processing: positive r2g, Top 10 region-to-gene links per gene: 2%| | 10/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 2%| | 13/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 3%| | 16/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 3%| | 19/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 4%| | 22/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 5%| | 26/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 5%| | 29/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 6%| | 32/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 6%| | 35/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 7%| | 38/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 7%| | 41/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 8%| | 44/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 8%| | 47/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 9%| | 51/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 9%| | 54/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 10%| | 57/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 10%| | 60/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 11%| | 63/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 11%| | 66/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 12%| | 69/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 13%|▏| 72/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 13%|▏| 75/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 14%|▏| 79/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 14%|▏| 82/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 15%|▏| 85/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 15%|▏| 88/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 16%|▏| 92/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 17%|▏| 96/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 17%|▏| 99/574 [
Processing: positive r2g, Top 10 region-to-gene links per gene: 18%|▏| 102/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 18%|▏| 106/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 19%|▏| 110/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 20%|▏| 113/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 20%|▏| 116/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 21%|▏| 119/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 21%|▏| 122/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 22%|▏| 125/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 22%|▏| 128/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 23%|▏| 131/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 23%|▏| 134/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 24%|▏| 138/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 25%|▏| 141/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 25%|▎| 144/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 26%|▎| 147/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 26%|▎| 150/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 27%|▎| 153/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 27%|▎| 156/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 28%|▎| 159/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 28%|▎| 162/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 29%|▎| 165/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 29%|▎| 168/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 30%|▎| 171/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 30%|▎| 174/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 31%|▎| 177/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 31%|▎| 180/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 32%|▎| 184/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 33%|▎| 187/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 33%|▎| 190/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 34%|▎| 193/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 34%|▎| 196/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 35%|▎| 199/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 35%|▎| 202/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 36%|▎| 205/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 36%|▎| 208/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 37%|▎| 212/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 37%|▎| 215/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 38%|▍| 218/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 39%|▍| 221/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 39%|▍| 224/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 40%|▍| 228/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 40%|▍| 231/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 41%|▍| 234/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 41%|▍| 238/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 42%|▍| 241/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 43%|▍| 244/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 43%|▍| 248/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 44%|▍| 251/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 44%|▍| 254/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 45%|▍| 257/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 45%|▍| 261/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 46%|▍| 264/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 47%|▍| 267/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 47%|▍| 270/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 48%|▍| 273/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 48%|▍| 276/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 49%|▍| 279/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 49%|▍| 283/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 50%|▍| 286/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 50%|▌| 289/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 51%|▌| 293/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 52%|▌| 297/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 52%|▌| 300/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 53%|▌| 303/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 53%|▌| 306/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 54%|▌| 309/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 54%|▌| 312/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 55%|▌| 315/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 55%|▌| 318/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 56%|▌| 321/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 56%|▌| 324/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 57%|▌| 327/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 57%|▌| 330/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 58%|▌| 333/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 59%|▌| 336/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 59%|▌| 339/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 60%|▌| 342/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 60%|▌| 346/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 61%|▌| 349/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 61%|▌| 352/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 62%|▌| 356/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 63%|▋| 359/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 63%|▋| 362/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 64%|▋| 365/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 64%|▋| 368/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 65%|▋| 371/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 65%|▋| 374/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 66%|▋| 377/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 66%|▋| 380/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 67%|▋| 383/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 67%|▋| 386/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 68%|▋| 389/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 68%|▋| 392/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 69%|▋| 395/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 69%|▋| 398/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 70%|▋| 401/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 71%|▋| 405/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 71%|▋| 408/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 72%|▋| 411/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 72%|▋| 414/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 73%|▋| 417/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 73%|▋| 420/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 74%|▋| 423/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 74%|▋| 426/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 75%|▋| 429/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 75%|▊| 432/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 76%|▊| 435/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 76%|▊| 438/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 77%|▊| 441/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 77%|▊| 444/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 78%|▊| 447/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 78%|▊| 450/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 79%|▊| 453/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 79%|▊| 456/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 80%|▊| 459/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 81%|▊| 463/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 81%|▊| 466/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 82%|▊| 470/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 82%|▊| 473/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 83%|▊| 476/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 83%|▊| 479/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 84%|▊| 482/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 84%|▊| 485/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 85%|▊| 489/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 86%|▊| 493/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 86%|▊| 496/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 87%|▊| 499/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 87%|▊| 502/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 88%|▉| 506/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 89%|▉| 509/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 89%|▉| 512/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 90%|▉| 515/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 90%|▉| 518/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 91%|▉| 521/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 91%|▉| 524/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 92%|▉| 528/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 93%|▉| 531/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 93%|▉| 534/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 94%|▉| 537/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 94%|▉| 540/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 95%|▉| 543/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 95%|▉| 546/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 96%|▉| 550/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 96%|▉| 553/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 97%|▉| 557/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 98%|▉| 561/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 98%|▉| 564/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 99%|▉| 567/574
Processing: positive r2g, Top 10 region-to-gene links per gene: 99%|▉| 571/574
86%|████████████████████████████████████▊ | 12/14 [02:52<00:32, 16.30s/it]
Processing: Top 15 region-to-gene links per gene, positive r2g: 0%| | 0/574 [0
Processing: Top 15 region-to-gene links per gene, positive r2g: 0%| | 2/574 [0
Processing: Top 15 region-to-gene links per gene, positive r2g: 1%| | 4/574 [0
Processing: Top 15 region-to-gene links per gene, positive r2g: 1%| | 6/574 [0
Processing: Top 15 region-to-gene links per gene, positive r2g: 2%| | 9/574 [0
Processing: Top 15 region-to-gene links per gene, positive r2g: 2%| | 11/574 [
Processing: Top 15 region-to-gene links per gene, positive r2g: 2%| | 13/574 [
Processing: Top 15 region-to-gene links per gene, positive r2g: 3%| | 15/574 [
Processing: Top 15 region-to-gene links per gene, positive r2g: 3%| | 17/574 [
Processing: Top 15 region-to-gene links per gene, positive r2g: 3%| | 19/574 [
Processing: Top 15 region-to-gene links per gene, positive r2g: 4%| | 21/574 [
Processing: Top 15 region-to-gene links per gene, positive r2g: 4%| | 23/574 [
Processing: Top 15 region-to-gene links per gene, positive r2g: 4%| | 25/574 [
Processing: Top 15 region-to-gene links per gene, positive r2g: 5%| | 27/574 [
Processing: Top 15 region-to-gene links per gene, positive r2g: 5%| | 30/574 [
Processing: Top 15 region-to-gene links per gene, positive r2g: 6%| | 33/574 [
Processing: Top 15 region-to-gene links per gene, positive r2g: 6%| | 35/574 [
Processing: Top 15 region-to-gene links per gene, positive r2g: 6%| | 37/574 [
Processing: Top 15 region-to-gene links per gene, positive r2g: 7%| | 39/574 [
Processing: Top 15 region-to-gene links per gene, positive r2g: 7%| | 41/574 [
Processing: Top 15 region-to-gene links per gene, positive r2g: 7%| | 43/574 [
Processing: Top 15 region-to-gene links per gene, positive r2g: 8%| | 45/574 [
100%|████████████████████████████████████████| 574/574 [00:05<00:00, 113.17it/s]
100%|███████████████████████████████████████| 329/329 [00:00<00:00, 1686.35it/s]
Running for Positive TF to gene: 0%| | 0/8001 [00:00<?, ?it/s]/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/gseapy/algorithm.py:71: RuntimeWarning: divide by zero encountered in divide
norm_tag = 1.0/sum_correl_tag
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/gseapy/algorithm.py:74: RuntimeWarning: invalid value encountered in multiply
RES = np.cumsum(tag_indicator * correl_vector * norm_tag - no_tag_indicator * norm_no_tag, axis=axis)
Running for Positive TF to gene: 0%| | 40/8001 [00:05<17:35, 7.54it/s]/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/gseapy/algorithm.py:71: RuntimeWarning: divide by zero encountered in divide
norm_tag = 1.0/sum_correl_tag
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/gseapy/algorithm.py:74: RuntimeWarning: invalid value encountered in multiply
RES = np.cumsum(tag_indicator * correl_vector * norm_tag - no_tag_indicator * norm_no_tag, axis=axis)
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/joblib/externals/loky/process_executor.py:752: UserWarning: A worker stopped while some jobs were given to the executor. This can be caused by a too short worker timeout or by a memory leak.
warnings.warn(
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/gseapy/algorithm.py:71: RuntimeWarning: divide by zero encountered in divide
norm_tag = 1.0/sum_correl_tag
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/gseapy/algorithm.py:74: RuntimeWarning: invalid value encountered in multiply
RES = np.cumsum(tag_indicator * correl_vector * norm_tag - no_tag_indicator * norm_no_tag, axis=axis)
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/gseapy/algorithm.py:71: RuntimeWarning: divide by zero encountered in divide
norm_tag = 1.0/sum_correl_tag
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/gseapy/algorithm.py:74: RuntimeWarning: invalid value encountered in multiply
RES = np.cumsum(tag_indicator * correl_vector * norm_tag - no_tag_indicator * norm_no_tag, axis=axis)
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/gseapy/algorithm.py:71: RuntimeWarning: divide by zero encountered in divide
norm_tag = 1.0/sum_correl_tag
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/gseapy/algorithm.py:74: RuntimeWarning: invalid value encountered in multiply
RES = np.cumsum(tag_indicator * correl_vector * norm_tag - no_tag_indicator * norm_no_tag, axis=axis)
Running for Positive TF to gene: 6%|▎ | 480/8001 [00:26<00:59, 125.90it/s]/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/gseapy/algorithm.py:71: RuntimeWarning: divide by zero encountered in divide
norm_tag = 1.0/sum_correl_tag
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/gseapy/algorithm.py:74: RuntimeWarning: invalid value encountered in multiply
RES = np.cumsum(tag_indicator * correl_vector * norm_tag - no_tag_indicator * norm_no_tag, axis=axis)
Running for Positive TF to gene: 100%|██████| 8001/8001 [01:23<00:00, 95.25it/s]
Running for Negative TF to gene: 100%|█████| 8001/8001 [00:24<00:00, 324.33it/s]
2024-03-11 16:07:34,786 GSEA INFO Subsetting on adjusted pvalue: 1, minimal NES: 0 and minimal leading edge genes 10
2024-03-11 16:07:34,868 GSEA INFO Merging eRegulons
2024-03-11 16:07:38,490 SCENIC+ INFO Formatting eGRN as table.
2024-03-11 16:07:39,407 SCENIC+ INFO Calculating triplet ranking.
2024-03-11 16:07:39,554 cisTarget INFO Reading cisTarget database
2024-03-11 16:08:19,063 SCENIC+ INFO Saving network to /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/eRegulon_direct.tsv
[Mon Mar 11 16:08:23 2024]
Finished job 4.
8 of 13 steps (62%) done
Select jobs to execute...
Execute 1 jobs...
[Mon Mar 11 16:08:23 2024]
localrule eGRN_extended:
input: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/tf_to_gene_adj.tsv, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/region_to_gene_adj.tsv, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/cistromes_extended.h5ad, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/ctx_db/10x_brain_1kb_bg_with_mask.regions_vs_motifs.rankings.feather
output: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/eRegulons_extended.tsv
jobid: 13
reason: Missing output files: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/eRegulons_extended.tsv; Input files updated by another job: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/region_to_gene_adj.tsv, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/cistromes_extended.h5ad, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/tf_to_gene_adj.tsv
threads: 20
resources: tmpdir=/tmp
2024-03-11 16:08:28,050 SCENIC+ INFO Loading TF to gene adjacencies.
2024-03-11 16:08:29,364 SCENIC+ INFO Loading region to gene adjacencies.
2024-03-11 16:08:31,454 SCENIC+ INFO Loading cistromes.
2024-03-11 16:08:31,505 GSEA INFO Thresholding region to gene relationships
0%| | 0/14 [00:00<?, ?it/s]
Processing: negative r2g, 0.85 quantile: 0%| | 0/318 [00:00<?, ?it/s]
Processing: negative r2g, 0.85 quantile: 2%| | 7/318 [00:00<00:04, 68.79it/s]
Processing: negative r2g, 0.85 quantile: 5%| | 17/318 [00:00<00:03, 86.11it/s]
Processing: negative r2g, 0.85 quantile: 8%| | 27/318 [00:00<00:03, 86.22it/s]
Processing: negative r2g, 0.85 quantile: 11%| | 36/318 [00:00<00:03, 81.99it/s]
Processing: negative r2g, 0.85 quantile: 14%|▏| 46/318 [00:00<00:03, 86.15it/s]
Processing: negative r2g, 0.85 quantile: 17%|▏| 55/318 [00:00<00:03, 84.72it/s]
Processing: negative r2g, 0.85 quantile: 20%|▏| 64/318 [00:00<00:02, 85.44it/s]
Processing: negative r2g, 0.85 quantile: 24%|▏| 75/318 [00:00<00:02, 91.01it/s]
Processing: negative r2g, 0.85 quantile: 27%|▎| 85/318 [00:01<00:04, 53.00it/s]
Processing: negative r2g, 0.85 quantile: 30%|▎| 94/318 [00:01<00:03, 59.57it/s]
Processing: negative r2g, 0.85 quantile: 32%|▎| 102/318 [00:01<00:03, 63.61it/s
Processing: negative r2g, 0.85 quantile: 35%|▎| 112/318 [00:01<00:02, 71.76it/s
Processing: negative r2g, 0.85 quantile: 38%|▍| 122/318 [00:01<00:02, 76.44it/s
Processing: negative r2g, 0.85 quantile: 41%|▍| 131/318 [00:01<00:02, 76.89it/s
Processing: negative r2g, 0.85 quantile: 45%|▍| 142/318 [00:01<00:02, 83.49it/s
Processing: negative r2g, 0.85 quantile: 47%|▍| 151/318 [00:01<00:01, 83.68it/s
Processing: negative r2g, 0.85 quantile: 50%|▌| 160/318 [00:02<00:02, 78.89it/s
Processing: negative r2g, 0.85 quantile: 54%|▌| 171/318 [00:02<00:01, 85.10it/s
Processing: negative r2g, 0.85 quantile: 57%|▌| 182/318 [00:02<00:01, 90.26it/s
Processing: negative r2g, 0.85 quantile: 60%|▌| 192/318 [00:02<00:01, 88.32it/s
Processing: negative r2g, 0.85 quantile: 63%|▋| 201/318 [00:02<00:02, 50.72it/s
Processing: negative r2g, 0.85 quantile: 66%|▋| 210/318 [00:02<00:01, 57.55it/s
Processing: negative r2g, 0.85 quantile: 69%|▋| 219/318 [00:03<00:01, 63.66it/s
Processing: negative r2g, 0.85 quantile: 71%|▋| 227/318 [00:03<00:01, 64.60it/s
Processing: negative r2g, 0.85 quantile: 74%|▋| 235/318 [00:03<00:01, 68.09it/s
Processing: negative r2g, 0.85 quantile: 76%|▊| 243/318 [00:03<00:01, 70.64it/s
Processing: negative r2g, 0.85 quantile: 79%|▊| 251/318 [00:03<00:01, 65.74it/s
Processing: negative r2g, 0.85 quantile: 81%|▊| 259/318 [00:03<00:00, 63.65it/s
Processing: negative r2g, 0.85 quantile: 84%|▊| 266/318 [00:03<00:00, 61.73it/s
Processing: negative r2g, 0.85 quantile: 86%|▊| 273/318 [00:03<00:00, 61.03it/s
Processing: negative r2g, 0.85 quantile: 88%|▉| 280/318 [00:03<00:00, 61.80it/s
Processing: negative r2g, 0.85 quantile: 91%|▉| 290/318 [00:04<00:00, 71.02it/s
Processing: negative r2g, 0.85 quantile: 95%|▉| 302/318 [00:04<00:00, 82.27it/s
Processing: negative r2g, 0.85 quantile: 98%|▉| 311/318 [00:04<00:00, 54.84it/s
7%|███▏ | 1/14 [00:06<01:29, 6.86s/it]
Processing: negative r2g, 0.9 quantile: 0%| | 0/318 [00:00<?, ?it/s]
Processing: negative r2g, 0.9 quantile: 4%| | 14/318 [00:00<00:02, 131.48it/s]
Processing: negative r2g, 0.9 quantile: 9%| | 28/318 [00:00<00:02, 124.45it/s]
Processing: negative r2g, 0.9 quantile: 13%|▎ | 41/318 [00:00<00:02, 97.56it/s]
Processing: negative r2g, 0.9 quantile: 16%|▏| 52/318 [00:00<00:02, 101.30it/s]
Processing: negative r2g, 0.9 quantile: 21%|▏| 67/318 [00:00<00:02, 114.41it/s]
Processing: negative r2g, 0.9 quantile: 25%|▏| 79/318 [00:00<00:02, 115.66it/s]
Processing: negative r2g, 0.9 quantile: 29%|▎| 93/318 [00:00<00:01, 120.33it/s]
Processing: negative r2g, 0.9 quantile: 33%|▎| 106/318 [00:00<00:01, 113.02it/s
Processing: negative r2g, 0.9 quantile: 37%|▎| 118/318 [00:01<00:01, 102.16it/s
Processing: negative r2g, 0.9 quantile: 41%|▍| 129/318 [00:01<00:02, 92.49it/s]
Processing: negative r2g, 0.9 quantile: 44%|▍| 141/318 [00:01<00:01, 98.27it/s]
Processing: negative r2g, 0.9 quantile: 48%|▍| 154/318 [00:01<00:01, 104.34it/s
Processing: negative r2g, 0.9 quantile: 52%|▌| 166/318 [00:01<00:01, 106.60it/s
Processing: negative r2g, 0.9 quantile: 56%|▌| 177/318 [00:01<00:01, 90.70it/s]
Processing: negative r2g, 0.9 quantile: 59%|▌| 189/318 [00:01<00:01, 96.78it/s]
Processing: negative r2g, 0.9 quantile: 63%|▋| 200/318 [00:01<00:01, 96.56it/s]
Processing: negative r2g, 0.9 quantile: 66%|▋| 210/318 [00:02<00:01, 97.38it/s]
Processing: negative r2g, 0.9 quantile: 70%|▋| 223/318 [00:02<00:00, 103.55it/s
Processing: negative r2g, 0.9 quantile: 74%|▋| 235/318 [00:02<00:01, 59.42it/s]
Processing: negative r2g, 0.9 quantile: 77%|▊| 245/318 [00:02<00:01, 65.82it/s]
Processing: negative r2g, 0.9 quantile: 80%|▊| 255/318 [00:02<00:00, 72.13it/s]
Processing: negative r2g, 0.9 quantile: 84%|▊| 266/318 [00:02<00:00, 79.82it/s]
Processing: negative r2g, 0.9 quantile: 87%|▊| 277/318 [00:02<00:00, 86.34it/s]
Processing: negative r2g, 0.9 quantile: 91%|▉| 289/318 [00:03<00:00, 94.00it/s]
Processing: negative r2g, 0.9 quantile: 95%|▉| 302/318 [00:03<00:00, 103.29it/s
Processing: negative r2g, 0.9 quantile: 99%|▉| 315/318 [00:03<00:00, 109.27it/s
14%|██████▎ | 2/14 [00:10<01:02, 5.20s/it]
Processing: 0.95 quantile, negative r2g: 0%| | 0/318 [00:00<?, ?it/s]
Processing: 0.95 quantile, negative r2g: 7%| | 21/318 [00:00<00:01, 203.13it/s
Processing: 0.95 quantile, negative r2g: 13%|▏| 42/318 [00:00<00:01, 198.27it/s
Processing: 0.95 quantile, negative r2g: 19%|▏| 62/318 [00:00<00:01, 199.02it/s
Processing: 0.95 quantile, negative r2g: 26%|▎| 83/318 [00:00<00:01, 197.57it/s
Processing: 0.95 quantile, negative r2g: 33%|▎| 106/318 [00:00<00:01, 205.79it/
Processing: 0.95 quantile, negative r2g: 40%|▍| 127/318 [00:00<00:00, 196.97it/
Processing: 0.95 quantile, negative r2g: 47%|▍| 148/318 [00:00<00:00, 198.47it/
Processing: 0.95 quantile, negative r2g: 53%|▌| 168/318 [00:00<00:00, 194.17it/
Processing: 0.95 quantile, negative r2g: 59%|▌| 188/318 [00:00<00:00, 188.18it/
Processing: 0.95 quantile, negative r2g: 65%|▋| 207/318 [00:01<00:00, 177.54it/
Processing: 0.95 quantile, negative r2g: 71%|▋| 227/318 [00:01<00:00, 183.00it/
Processing: 0.95 quantile, negative r2g: 77%|▊| 246/318 [00:01<00:00, 172.97it/
Processing: 0.95 quantile, negative r2g: 83%|▊| 264/318 [00:01<00:00, 95.16it/s
Processing: 0.95 quantile, negative r2g: 89%|▉| 284/318 [00:01<00:00, 113.52it/
Processing: 0.95 quantile, negative r2g: 96%|▉| 306/318 [00:01<00:00, 134.14it/
21%|█████████▍ | 3/14 [00:13<00:44, 4.06s/it]
Processing: negative r2g, Top 5 region-to-gene links per gene: 0%| | 0/318 [00
Processing: negative r2g, Top 5 region-to-gene links per gene: 2%| | 7/318 [00
Processing: negative r2g, Top 5 region-to-gene links per gene: 4%| | 14/318 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 7%| | 21/318 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 9%| | 29/318 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 12%| | 37/318 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 14%|▏| 45/318 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 16%|▏| 52/318 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 19%|▏| 61/318 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 21%|▏| 68/318 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 24%|▏| 75/318 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 26%|▎| 83/318 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 29%|▎| 92/318 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 31%|▎| 99/318 [0
Processing: negative r2g, Top 5 region-to-gene links per gene: 33%|▎| 106/318 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 36%|▎| 113/318 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 38%|▍| 120/318 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 40%|▍| 127/318 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 42%|▍| 134/318 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 44%|▍| 141/318 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 47%|▍| 149/318 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 49%|▍| 157/318 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 52%|▌| 164/318 [
Processing: negative r2g, Top 5 region-to-gene links per gene: 54%|▌| 171/318 [
Running for Positive TF to gene: 100%|██████| 4431/4431 [00:56<00:00, 77.97it/s]
Running for Negative TF to gene: 100%|█████| 4431/4431 [00:12<00:00, 350.83it/s]
2024-03-11 16:12:15,195 GSEA INFO Subsetting on adjusted pvalue: 1, minimal NES: 0 and minimal leading edge genes 10
2024-03-11 16:12:15,256 GSEA INFO Merging eRegulons
2024-03-11 16:12:16,866 SCENIC+ INFO Formatting eGRN as table.
2024-03-11 16:12:17,774 SCENIC+ INFO Calculating triplet ranking.
2024-03-11 16:12:17,876 cisTarget INFO Reading cisTarget database
2024-03-11 16:13:05,393 SCENIC+ INFO Saving network to /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/eRegulons_extended.tsv
[Mon Mar 11 16:13:08 2024]
Finished job 13.
9 of 13 steps (69%) done
Select jobs to execute...
Execute 1 jobs...
[Mon Mar 11 16:13:08 2024]
localrule AUCell_direct:
input: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/eRegulon_direct.tsv, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/ACC_GEX.h5mu
output: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/AUCell_direct.h5mu
jobid: 3
reason: Missing output files: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/AUCell_direct.h5mu; Input files updated by another job: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/eRegulon_direct.tsv
threads: 20
resources: tmpdir=/tmp
2024-03-11 16:13:15,171 SCENIC+ INFO Reading data.
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
2024-03-11 16:13:18,838 SCENIC+ INFO Calculating enrichment scores.
2024-03-11 16:14:10,146 SCENIC+ INFO Writing file to /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/AUCell_direct.h5mu
[Mon Mar 11 16:14:10 2024]
Finished job 3.
10 of 13 steps (77%) done
Select jobs to execute...
Execute 1 jobs...
[Mon Mar 11 16:14:10 2024]
localrule AUCell_extended:
input: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/eRegulons_extended.tsv, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/ACC_GEX.h5mu
output: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/AUCell_extended.h5mu
jobid: 12
reason: Missing output files: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/AUCell_extended.h5mu; Input files updated by another job: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/eRegulons_extended.tsv
threads: 20
resources: tmpdir=/tmp
2024-03-11 16:14:15,853 SCENIC+ INFO Reading data.
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
2024-03-11 16:14:18,525 SCENIC+ INFO Calculating enrichment scores.
2024-03-11 16:15:09,670 SCENIC+ INFO Writing file to /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/AUCell_extended.h5mu
[Mon Mar 11 16:15:10 2024]
Finished job 12.
11 of 13 steps (85%) done
Select jobs to execute...
Execute 1 jobs...
[Mon Mar 11 16:15:10 2024]
localrule scplus_mudata:
input: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/ACC_GEX.h5mu, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/AUCell_direct.h5mu, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/AUCell_extended.h5mu, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/eRegulon_direct.tsv, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/eRegulons_extended.tsv
output: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/scplusmdata.h5mu
jobid: 1
reason: Missing output files: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/scplusmdata.h5mu; Input files updated by another job: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/eRegulon_direct.tsv, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/AUCell_extended.h5mu, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/AUCell_direct.h5mu, /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/eRegulons_extended.tsv
resources: tmpdir=/tmp
2024-03-11 16:15:15,108 SCENIC+ INFO Reading multiome MuData.
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
2024-03-11 16:15:17,756 SCENIC+ INFO Reading AUC values.
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
2024-03-11 16:15:17,815 SCENIC+ INFO Reading eRegulon metadata.
2024-03-11 16:15:17,841 SCENIC+ INFO Generating MuData object.
2024-03-11 16:15:18,699 SCENIC+ INFO Writing file to /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/scplusmdata.h5mu
[Mon Mar 11 16:15:23 2024]
Finished job 1.
12 of 13 steps (92%) done
Select jobs to execute...
Execute 1 jobs...
[Mon Mar 11 16:15:23 2024]
localrule all:
input: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/scplusmdata.h5mu
jobid: 0
reason: Input files updated by another job: /staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial/outs/scplusmdata.h5mu
resources: tmpdir=/tmp
[Mon Mar 11 16:15:23 2024]
Finished job 0.
13 of 13 steps (100%) done
Complete log: .snakemake/log/2024-03-11T151644.790329.snakemake.log
Main outputs#
The main output of the pipeline is the scplusmdata.h5mu file. This is a MuData file containing the eRegulons and enrichment scores.
[125]:
import os
os.chdir("/staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial")
[126]:
import mudata
scplus_mdata = mudata.read("outs/scplusmdata.h5mu")
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
Direct and extended predicted TF-to-region-to-gene links. This dataframe contains also a ranking of each TF-region-gene triplet, based on its importance triplet_rank.
[127]:
scplus_mdata.uns["direct_e_regulon_metadata"]
[127]:
| Region | Gene | importance_R2G | rho_R2G | importance_x_rho | importance_x_abs_rho | TF | is_extended | eRegulon_name | Gene_signature_name | Region_signature_name | importance_TF2G | regulation | rho_TF2G | triplet_rank | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | chr20:35305658-35306158 | CEP250 | 0.020002 | 0.056479 | 0.001130 | 0.001130 | BCL11A | False | BCL11A_direct_+/+ | BCL11A_direct_+/+_(352g) | BCL11A_direct_+/+_(641r) | 0.743348 | 1 | 0.149413 | 1879 |
| 1 | chr4:41408947-41409447 | UCHL1 | 0.039919 | 0.278051 | 0.011099 | 0.011099 | BCL11A | False | BCL11A_direct_+/+ | BCL11A_direct_+/+_(352g) | BCL11A_direct_+/+_(641r) | 1.572051 | 1 | 0.309145 | 1767 |
| 2 | chr10:133327408-133327908 | CALY | 0.021935 | 0.542038 | 0.011889 | 0.011889 | BCL11A | False | BCL11A_direct_+/+ | BCL11A_direct_+/+_(352g) | BCL11A_direct_+/+_(641r) | 0.934755 | 1 | 0.369021 | 7337 |
| 3 | chr1:239814303-239814803 | CHRM3 | 0.054480 | 0.731600 | 0.039857 | 0.039857 | BCL11A | False | BCL11A_direct_+/+ | BCL11A_direct_+/+_(352g) | BCL11A_direct_+/+_(641r) | 1.682106 | 1 | 0.497771 | 4406 |
| 4 | chr10:13987335-13987835 | FRMD4A | 0.002173 | 0.372226 | 0.000809 | 0.000809 | BCL11A | False | BCL11A_direct_+/+ | BCL11A_direct_+/+_(352g) | BCL11A_direct_+/+_(641r) | 0.764116 | 1 | 0.411129 | 6392 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 7648 | chr12:78102733-78103233 | NAV3 | 0.061226 | -0.598159 | -0.036623 | 0.036623 | TCF12 | False | TCF12_direct_-/- | TCF12_direct_-/-_(123g) | TCF12_direct_-/-_(136r) | 5.374822 | -1 | -0.474900 | 791 |
| 7649 | chr18:36376597-36377097 | FHOD3 | 0.006648 | -0.060058 | -0.000399 | 0.000399 | TCF12 | False | TCF12_direct_-/- | TCF12_direct_-/-_(123g) | TCF12_direct_-/-_(136r) | 1.216466 | -1 | -0.411142 | 4699 |
| 7650 | chr14:77307021-77307521 | TMED8 | 0.011415 | -0.258848 | -0.002955 | 0.002955 | TCF12 | False | TCF12_direct_-/- | TCF12_direct_-/-_(123g) | TCF12_direct_-/-_(136r) | 1.879806 | -1 | -0.217667 | 6561 |
| 7651 | chr14:103019978-103020478 | TRAF3 | 0.035434 | -0.251028 | -0.008895 | 0.008895 | TCF12 | False | TCF12_direct_-/- | TCF12_direct_-/-_(123g) | TCF12_direct_-/-_(136r) | 1.481711 | -1 | -0.183403 | 5162 |
| 7652 | chr12:20587950-20588450 | PDE3A | 0.018121 | -0.314249 | -0.005695 | 0.005695 | TCF12 | False | TCF12_direct_-/- | TCF12_direct_-/-_(123g) | TCF12_direct_-/-_(136r) | 2.102950 | -1 | -0.393440 | 2064 |
7653 rows × 15 columns
[128]:
scplus_mdata.uns["extended_e_regulon_metadata"]
[128]:
| Region | Gene | importance_R2G | rho_R2G | importance_x_rho | importance_x_abs_rho | TF | is_extended | eRegulon_name | Gene_signature_name | Region_signature_name | importance_TF2G | regulation | rho_TF2G | triplet_rank | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | chr1:6475533-6476033 | TNFRSF25 | 0.019441 | 0.217651 | 0.004231 | 0.004231 | EGR3 | True | EGR3_extended_+/+ | EGR3_extended_+/+_(24g) | EGR3_extended_+/+_(30r) | 1.070784 | 1 | 0.231038 | 4058 |
| 1 | chr2:241803041-241803541 | ATG4B | 0.015792 | 0.066621 | 0.001052 | 0.001052 | EGR3 | True | EGR3_extended_+/+ | EGR3_extended_+/+_(24g) | EGR3_extended_+/+_(30r) | 1.234643 | 1 | 0.074444 | 5455 |
| 2 | chr2:42101928-42102428 | EML4 | 0.037126 | 0.276649 | 0.010271 | 0.010271 | EGR3 | True | EGR3_extended_+/+ | EGR3_extended_+/+_(24g) | EGR3_extended_+/+_(30r) | 1.235069 | 1 | 0.202865 | 2496 |
| 3 | chr10:114303963-114304463 | TDRD1 | 0.082349 | 0.078376 | 0.006454 | 0.006454 | EGR3 | True | EGR3_extended_+/+ | EGR3_extended_+/+_(24g) | EGR3_extended_+/+_(30r) | 0.865910 | 1 | 0.071393 | 1707 |
| 4 | chr19:18161129-18161629 | IFI30 | 0.027840 | 0.106714 | 0.002971 | 0.002971 | EGR3 | True | EGR3_extended_+/+ | EGR3_extended_+/+_(24g) | EGR3_extended_+/+_(30r) | 0.962073 | 1 | 0.354533 | 3209 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 6137 | chr22:25436989-25437489 | GRK3 | 0.003550 | -0.114036 | -0.000405 | 0.000405 | TCF12 | True | TCF12_extended_-/- | TCF12_extended_-/-_(174g) | TCF12_extended_-/-_(202r) | 1.927643 | -1 | -0.302335 | 4648 |
| 6138 | chr12:104340926-104341426 | CHST11 | 0.001517 | -0.152301 | -0.000231 | 0.000231 | TCF12 | True | TCF12_extended_-/- | TCF12_extended_-/-_(174g) | TCF12_extended_-/-_(202r) | 1.120303 | -1 | -0.326319 | 4314 |
| 6139 | chr20:41184836-41185336 | ZHX3 | 0.001790 | -0.133616 | -0.000239 | 0.000239 | TCF12 | True | TCF12_extended_-/- | TCF12_extended_-/-_(174g) | TCF12_extended_-/-_(202r) | 1.006590 | -1 | -0.296368 | 3888 |
| 6140 | chr5:66827771-66828271 | MAST4 | 0.002882 | -0.218997 | -0.000631 | 0.000631 | TCF12 | True | TCF12_extended_-/- | TCF12_extended_-/-_(174g) | TCF12_extended_-/-_(202r) | 2.580701 | -1 | -0.420271 | 2502 |
| 6141 | chr15:34337164-34337664 | GOLGA8A | 0.026024 | -0.375745 | -0.009779 | 0.009779 | TCF12 | True | TCF12_extended_-/- | TCF12_extended_-/-_(174g) | TCF12_extended_-/-_(202r) | 2.319901 | -1 | -0.286801 | 2936 |
6142 rows × 15 columns
Downstream analysis#
eRegulon dimensionality reduction#
The eRegulon enrichment scores can be used to perform dimensionality reductions
[130]:
import scanpy as sc
import anndata
eRegulon_gene_AUC = anndata.concat(
[scplus_mdata["direct_gene_based_AUC"], scplus_mdata["extended_gene_based_AUC"]],
axis = 1,
)
[131]:
eRegulon_gene_AUC.obs = scplus_mdata.obs.loc[eRegulon_gene_AUC.obs_names]
[132]:
sc.pp.neighbors(eRegulon_gene_AUC, use_rep = "X")
[133]:
sc.tl.umap(eRegulon_gene_AUC)
[134]:
sc.pl.umap(eRegulon_gene_AUC, color = "scRNA_counts:Seurat_cell_type")
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/scanpy/plotting/_tools/scatterplots.py:378: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
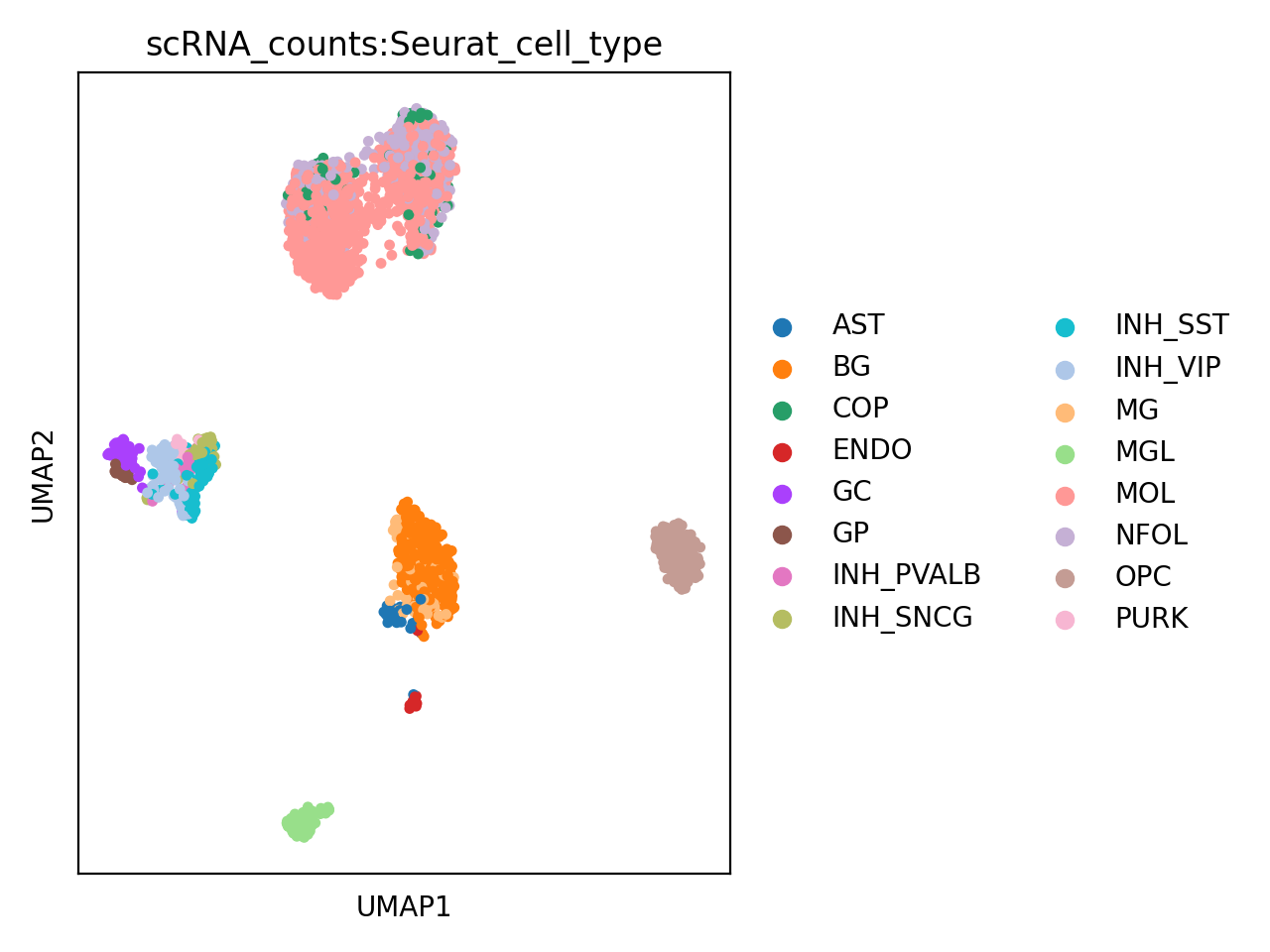
eRegulon specificity score#
[135]:
from scenicplus.RSS import (regulon_specificity_scores, plot_rss)
[136]:
rss = regulon_specificity_scores(
scplus_mudata = scplus_mdata,
variable = "scRNA_counts:Seurat_cell_type",
modalities = ["direct_gene_based_AUC", "extended_gene_based_AUC"]
)
[137]:
plot_rss(
data_matrix = rss,
top_n = 3,
num_columns = 5
)
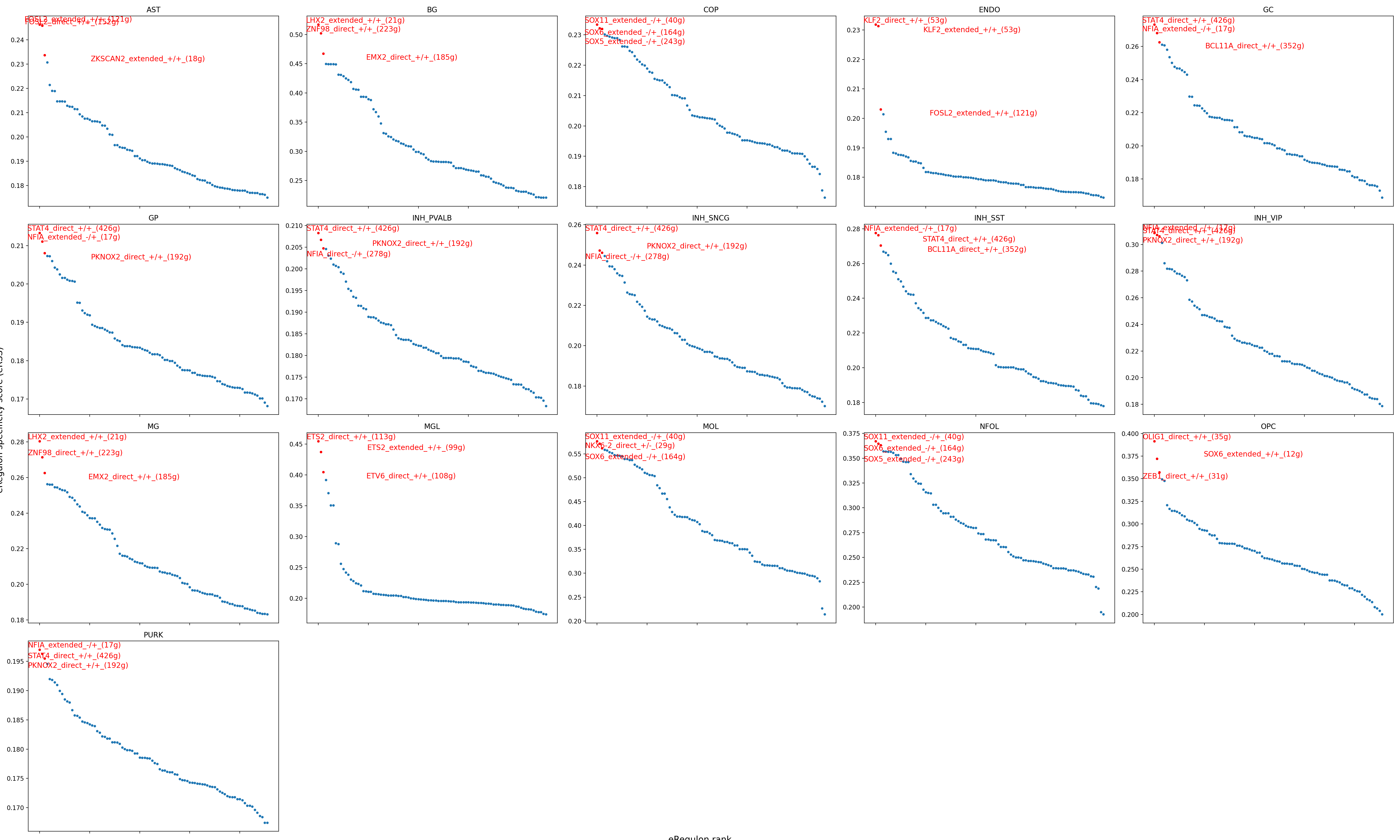
Plot eRegulon enrichment scores#
eRegulon enrichment scores can be plotted on the UMAP.
[138]:
sc.pl.umap(eRegulon_gene_AUC, color = list(set([x for xs in [rss.loc[ct].sort_values()[0:2].index for ct in rss.index] for x in xs ])))
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/IPython/core/pylabtools.py:152: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
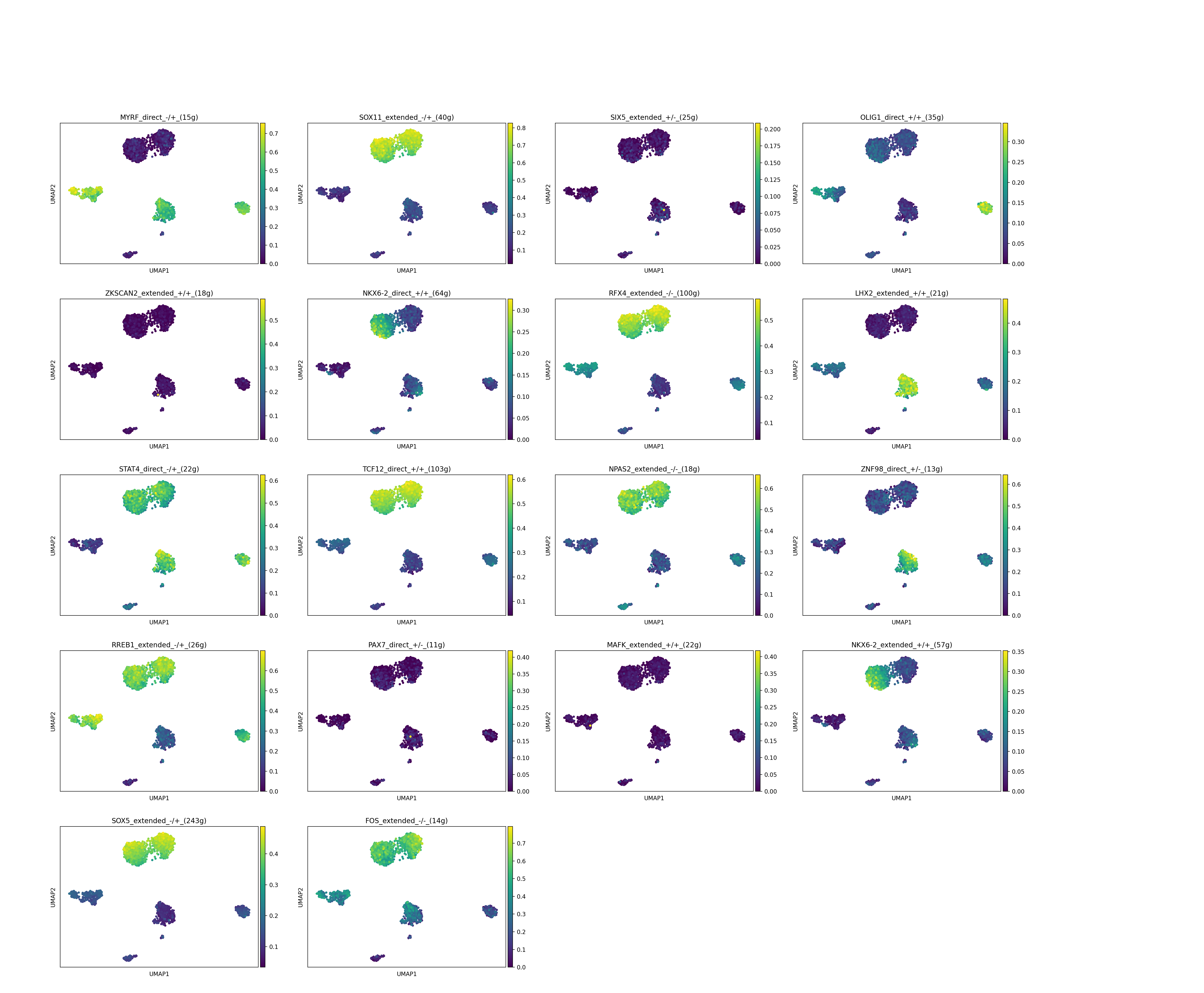
Heatmap dotplot#
We can draw a heatmap where the color represent target gene enrichment and the dotsize target region enrichment.
[142]:
from scenicplus.plotting.dotplot import heatmap_dotplot
[143]:
heatmap_dotplot(
scplus_mudata = scplus_mdata,
color_modality = "direct_gene_based_AUC",
size_modality = "direct_region_based_AUC",
group_variable = "scRNA_counts:Seurat_cell_type",
eRegulon_metadata_key = "direct_e_regulon_metadata",
color_feature_key = "Gene_signature_name",
size_feature_key = "Region_signature_name",
feature_name_key = "eRegulon_name",
sort_data_by = "direct_gene_based_AUC",
orientation = "horizontal",
figsize = (16, 5)
)
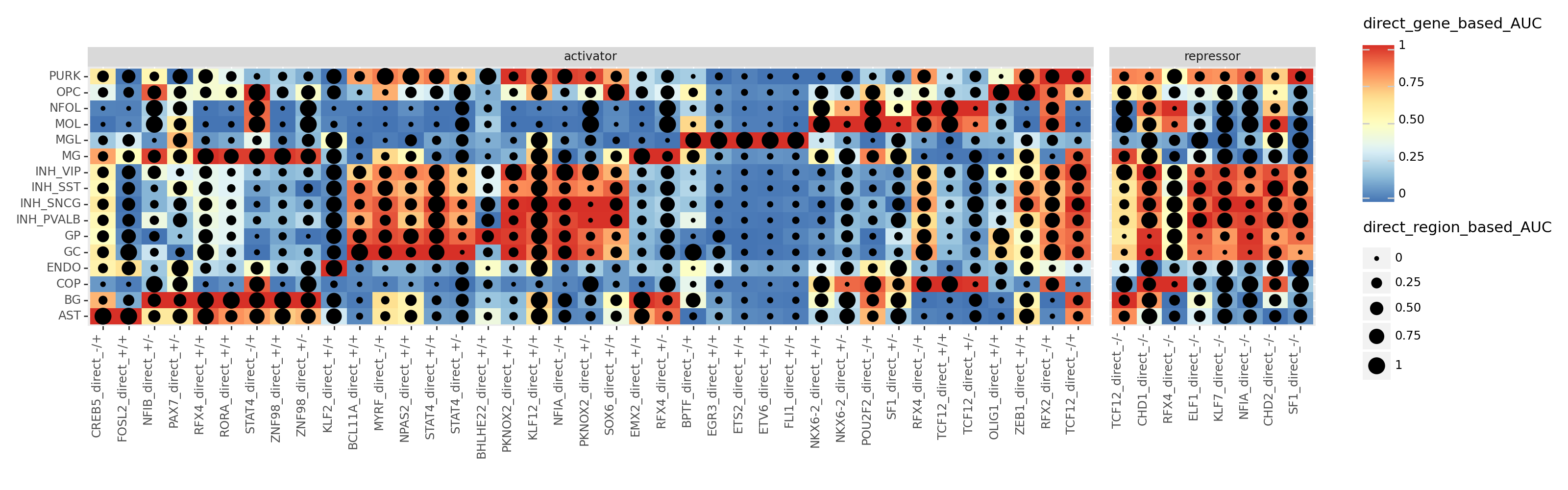
[143]:
<Figure Size: (1600 x 500)>
Converting mudata output to old-style SCENIC+ object.#
Not all functions in the original release of SCENIC+ are updated to use the new mudata output of SCENIC+. To be able to still use these old functions while they get updated we have a function to convert the mudata object the the old SCENIC+ object.
[1]:
import os
os.chdir("/staging/leuven/stg_00002/lcb/sdewin/PhD/python_modules/scenicplus_development_tutorial")
[2]:
import mudata
scplus_mdata = mudata.read("outs/scplusmdata.h5mu")
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
/lustre1/project/stg_00002/mambaforge/vsc33053/envs/scenicplus_development_tutorial/lib/python3.11/site-packages/anndata/_core/anndata.py:522: FutureWarning: The dtype argument is deprecated and will be removed in late 2024.
warnings.warn(
[3]:
from scenicplus.scenicplus_class import mudata_to_scenicplus
To regenerate the complete SCENIC+ object it is necessary to provide the paths to your motif enrichment results. However, this data is not necessary for most downstream functions so providing these paths is completely optional.
[4]:
scplus_obj = mudata_to_scenicplus(
mdata = scplus_mdata,
path_to_cistarget_h5 = "outs/ctx_results.hdf5",
path_to_dem_h5 = "outs/dem_results.hdf5"
)
[5]:
scplus_obj
[5]:
SCENIC+ object with n_cells x n_genes = 1970 x 36601 and n_cells x n_regions = 1970 x 435925
metadata_regions:'Chromosome', 'Start', 'End', 'Width', 'cisTopic_nr_frag', 'cisTopic_log_nr_frag', 'cisTopic_nr_acc', 'cisTopic_log_nr_acc'
metadata_genes:'gene_ids', 'feature_types'
metadata_cell:'VSN_cell_type', 'VSN_leiden_res0.3', 'VSN_leiden_res0.6', 'VSN_leiden_res0.9', 'VSN_leiden_res1.2', 'VSN_sample_id', 'Seurat_leiden_res0.6', 'Seurat_leiden_res1.2', 'Seurat_cell_type', 'n_genes_by_counts', 'total_counts', 'total_counts_mt', 'pct_counts_mt'
menr:'cistarget_DARs_cell_type_AST', 'cistarget_DARs_cell_type_BG', 'cistarget_DARs_cell_type_COP', 'cistarget_DARs_cell_type_ENDO', 'cistarget_DARs_cell_type_GC', 'cistarget_DARs_cell_type_GP', 'cistarget_DARs_cell_type_INH_PVALB', 'cistarget_DARs_cell_type_INH_SNCG', 'cistarget_DARs_cell_type_INH_SST', 'cistarget_DARs_cell_type_INH_VIP', 'cistarget_DARs_cell_type_MG', 'cistarget_DARs_cell_type_MGL', 'cistarget_DARs_cell_type_MOL', 'cistarget_DARs_cell_type_NFOL', 'cistarget_DARs_cell_type_OPC', 'cistarget_DARs_cell_type_PURK', 'cistarget_Topics_otsu_Topic1', 'cistarget_Topics_otsu_Topic10', 'cistarget_Topics_otsu_Topic11', 'cistarget_Topics_otsu_Topic12', 'cistarget_Topics_otsu_Topic13', 'cistarget_Topics_otsu_Topic14', 'cistarget_Topics_otsu_Topic15', 'cistarget_Topics_otsu_Topic16', 'cistarget_Topics_otsu_Topic17', 'cistarget_Topics_otsu_Topic18', 'cistarget_Topics_otsu_Topic19', 'cistarget_Topics_otsu_Topic2', 'cistarget_Topics_otsu_Topic20', 'cistarget_Topics_otsu_Topic21', 'cistarget_Topics_otsu_Topic22', 'cistarget_Topics_otsu_Topic23', 'cistarget_Topics_otsu_Topic24', 'cistarget_Topics_otsu_Topic25', 'cistarget_Topics_otsu_Topic26', 'cistarget_Topics_otsu_Topic27', 'cistarget_Topics_otsu_Topic28', 'cistarget_Topics_otsu_Topic29', 'cistarget_Topics_otsu_Topic3', 'cistarget_Topics_otsu_Topic30', 'cistarget_Topics_otsu_Topic31', 'cistarget_Topics_otsu_Topic32', 'cistarget_Topics_otsu_Topic33', 'cistarget_Topics_otsu_Topic34', 'cistarget_Topics_otsu_Topic35', 'cistarget_Topics_otsu_Topic36', 'cistarget_Topics_otsu_Topic37', 'cistarget_Topics_otsu_Topic38', 'cistarget_Topics_otsu_Topic39', 'cistarget_Topics_otsu_Topic4', 'cistarget_Topics_otsu_Topic40', 'cistarget_Topics_otsu_Topic5', 'cistarget_Topics_otsu_Topic6', 'cistarget_Topics_otsu_Topic7', 'cistarget_Topics_otsu_Topic8', 'cistarget_Topics_otsu_Topic9', 'cistarget_Topics_top_3k_Topic1', 'cistarget_Topics_top_3k_Topic10', 'cistarget_Topics_top_3k_Topic11', 'cistarget_Topics_top_3k_Topic12', 'cistarget_Topics_top_3k_Topic13', 'cistarget_Topics_top_3k_Topic14', 'cistarget_Topics_top_3k_Topic15', 'cistarget_Topics_top_3k_Topic16', 'cistarget_Topics_top_3k_Topic17', 'cistarget_Topics_top_3k_Topic18', 'cistarget_Topics_top_3k_Topic19', 'cistarget_Topics_top_3k_Topic2', 'cistarget_Topics_top_3k_Topic20', 'cistarget_Topics_top_3k_Topic21', 'cistarget_Topics_top_3k_Topic22', 'cistarget_Topics_top_3k_Topic23', 'cistarget_Topics_top_3k_Topic24', 'cistarget_Topics_top_3k_Topic25', 'cistarget_Topics_top_3k_Topic26', 'cistarget_Topics_top_3k_Topic27', 'cistarget_Topics_top_3k_Topic28', 'cistarget_Topics_top_3k_Topic29', 'cistarget_Topics_top_3k_Topic3', 'cistarget_Topics_top_3k_Topic30', 'cistarget_Topics_top_3k_Topic31', 'cistarget_Topics_top_3k_Topic32', 'cistarget_Topics_top_3k_Topic33', 'cistarget_Topics_top_3k_Topic34', 'cistarget_Topics_top_3k_Topic35', 'cistarget_Topics_top_3k_Topic36', 'cistarget_Topics_top_3k_Topic37', 'cistarget_Topics_top_3k_Topic38', 'cistarget_Topics_top_3k_Topic39', 'cistarget_Topics_top_3k_Topic4', 'cistarget_Topics_top_3k_Topic40', 'cistarget_Topics_top_3k_Topic5', 'cistarget_Topics_top_3k_Topic6', 'cistarget_Topics_top_3k_Topic7', 'cistarget_Topics_top_3k_Topic8', 'cistarget_Topics_top_3k_Topic9', 'dem_DARs_cell_type_INH_PVALB_vs_all', 'dem_DARs_cell_type_NFOL_vs_all', 'dem_Topics_otsu_Topic14_vs_all', 'dem_Topics_otsu_Topic26_vs_all', 'dem_Topics_otsu_Topic40_vs_all', 'dem_Topics_top_3k_Topic14_vs_all', 'dem_Topics_top_3k_Topic22_vs_all', 'dem_Topics_top_3k_Topic26_vs_all', 'dem_Topics_top_3k_Topic40_vs_all', 'dem_Topics_top_3k_Topic7_vs_all'
dr_cell:'rna_X_pca', 'rna_X_tsne', 'rna_X_umap'
